Cellular communication and function depend on potential energy. Cellular activities like action potentials, muscle contractions, and the control of voltage gated ion channels all depend on transmitting and sensing electrical potential. Thankfully, there is a tool to measure these electrical events! Voltage indicators are voltmeters for cells! In this blog we will review how these indicators work, their general applications, and why viruses are essential to their use as biological tools.
Why Use Voltage Indicators?
Electrical signals are integral to many facets of biology. For example, many heart issues stem from the heart beating at an irregular or insufficient level. The heartbeat – a muscle contraction – is directly related to the electrical impulse of the cardiac cells. To study this phenomenon you will likely want to monitor the voltage change across the membrane of cardiac cells. Similarly, neuronal signaling is carried out by action potentials which are electrical charges propagated through multiple cells to do things like translate perceived signals to physical actions. Understanding neuronal pathways and the function of certain cells depends on the ability to monitor their signals – electricity!
The research tools for studying voltage within a cell are genetically encoded voltage indicators (GEVIs) and chemigenetic sensors. These tools have two main elements: a voltage-sensing component and a fluorescent output. The GEVI family of gadgets includes Ace, JEDI, and many more. They can be targeted to the plasma membrane to measure membrane potential and can also be directed to specific types of cells, such as neuron subtypes, thanks to viral serotype specificity during delivery (more on this later). Chemigenetic sensors take a hybrid approach and utilize both a genetic component as well as a small molecule which we will describe in more detail below.
Types of Voltage Indicators
GEVIs can be divided up structurally into 2 main groups: voltage sensitive domains and Rhodopsin containing GEVIs. Chemigenetic sensors are better classified by their fluorescent reporter and the mechanism of their voltage sensing component, which separates them into two distinct categories detailed below.
Voltage Sensitive Domain (VSD) GEVIs
Simply put, VSD-based sensors consist of a VSD fused to a single fluorescent protein (FP). This class of GEVI was the earliest developed voltage sensor with a genetic component. The VSD components have historically come from a variety of sources – voltage-gated potassium channels found in Drosophila to phosphatases from sea squirts. This design structurally separates the two components of the indicator, which can make the kinetics of voltage sensing to signal output slightly slower than other approaches. Functionally, the voltage sensitivity of the available VSDs has been successful even at the single neuron level in vitro. In vivo monitoring has also been achieved with these sensors, but further optimization is necessary for these to be as effective at the single cell level as they are in vitro. Factors like photobleaching, lack of red-shifted indicators, and brightness of fluorescence all have room for improvement. These sensors still have many useful applications, though, and if you’re in the market for a VSD GEVI, the JEDI-2P is an excellent choice.
Rhodopsin GEVIs
Rhodopsin is composed of an opsin (a GPCR) linked to a chromophore and functions as a photoreceptor in the eye. There are two flavors of rhodopsin GEVIs. The first is a two-in-one system where rhodopsin functions as both the voltage detector and fluorescent signaler. The pro of this system is the kinetic response time from signal input to readable output, which generally exceeds that of the VSD GEVIs. However, since rhodopsin proteins were initially evolved for ion transport, not fluorescence, they are not nearly as bright as the other GEVI systems that use engineered FPs. Attempts have been made to optimize their fluorescence, which ultimately led to the engineering of the second type of these GEVIs: a FP-fused rhodopsin.
FP-fusion to rhodopsin resulted in electrochromic FRET capable GEVIs, with rhodopsin quenching the added fluor for this two-part fluorescent system, and rhodopsin still functioning as the voltage sensor. FP-rhodopsin GEVIs have been engineered to have kinetics comparable to its one component counterpart and is significantly brighter and available with red-shifted fluorophores (find FRET capable GEVIs here!). The only issue with these GEVIs? They perform poorly in two-photon conditions, which are necessary conditions for penetrating deeper tissues depths (1 mm), important for certain applications such as in vivo tissue imaging.
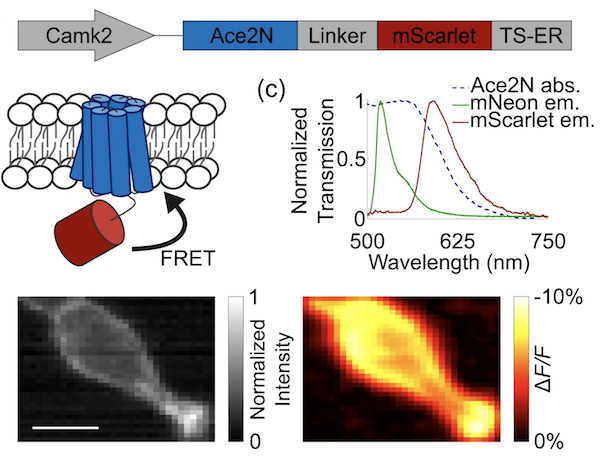
|
|
Figure 1: Ace-mScarlet is a fusion between the voltage-sensitive inhibitory rhodopsin Ace2N and mScarlet. Ace2N quenches a proportion of mScarlet's emission via FRET. Image from Beck et al, 2019 under CC BY 4.0. |
FRET Chemigenetic Sensors
These types of sensors can have a variety of different components, but all contain a FRET pair, a genetic component, and a small molecule component. Early versions of these sensors contained a FP that localized to the plasma membrane due to the addition of several targeting domains. The quencher and voltage detector in this system was dipicrylamine, a lipophilic compound that distributes itself within the plasma membrane as well.
Another flavor of these tools is Voltron, which uses rhodopsin as the voltage sensing component and a self-labeling domain that is capable of binding a Janelia Fluor (JF). The voltage-dependent changes in rhodopsin modulate the degree of quenching of the fluor via FRET. This system is particularly powerful for in vivo imaging especially since JFs can cross the blood brain barrier and many colors of JFs are available to swap into the system.
Single Fluor Chemigenetic Sensors
This system consists of a genetically expressed enzyme, typically on the cell surface, which can act on an upstream component, the chemical factor, of a naturally occurring voltage indicator. To give an example, a genetically encoded phosphatase (voltage sensing) hydrolyzes a precursor dye to make it more lipophilic, increasing its affinity for the membrane, where it will then localize and emit signal from. This specific example is attractive due to the large number of sheer molecules and signal you can generate without disturbing the integrity of the membrane, a concern with several of the previously described systems. Several adaptations of this method have been to tag, cage, uncage, and trap the chemical fluor with the genetically encoded component of the system to offer unique advantages depending on your experimental system.
%20(1).jpeg?width=710&height=328&name=Chemigenetics%20sensor%20diagram%20(1)%20(1).jpeg)
|
|
Figure 2: General overview of chemigenetic tools. A genetically encoded component is delivered in vivo or in vitro and a small molecule is added exogenously to measure a phenotypic response in the form of fluorescence or voltage indicators. |
Viral Vectors and Voltage Indicators
As previously mentioned, cellular voltage is measured at the cell membrane, and this voltage is particularly physiologically relevant in neurons, myocytes, and endocrine cells. It follows that the specific targeting of these voltage indicators needs to be cell type specific as well as specifically localize to the membrane, otherwise they aren’t all that useful. The cellular localization component is typically handled by either the fluorescent component (i.e., when the dye is lipophilic and integrates into the membrane), the voltage sensing component (i.e., rhodopsin naturally resides in the membrane), or both. Targeting these systems to specific cell types is where viral vectors become invaluable.
Viral vectors are mechanisms by which genetic components can be delivered to cells both transiently and for genomic integration. Lab-engineered viruses will package your genetic cargo in lieu of their own genomes and deposit this cargo upon host infection. Packaging viruses also have various serotypes – the surface proteins that confer affinity for a target – and this serotype can be selected to preferentially target your cell type of interest. Viral pseudotyping is also a powerful tool that swaps capsid proteins from different viruses to optimize them for targeting your cell type of interest. For in vivo imaging applications, the ability to direct your voltage sensor selectively is especially essential. If you want to learn more about how viruses can be used for chemigenetics, biosensors, and generally as biological tools, our blog has you covered. Happy voltage sensing!
-1.jpeg?width=841&height=336&name=Untitled%20(1)-1.jpeg)
|
| Figure 3: Components of the types of voltage indicators reviewed. |
References and Resources
References
Beck C, Zhang D, Gong Y. Enhanced genetically encoded voltage indicators advance their applications in neuroscience. Curr Opin Biomed Eng. 2019. 12:111-117. doi: 10.1016/j.cobme.2019.10.010.
Bando, Y., Grimm, C., Cornejo, V.H. et al. Genetic voltage indicators. BMC Biol 17, 71 (2019). doi:10.1186/s12915-019-0682-0.
Beck, C., Gong, Y. A high-speed, bright, red fluorescent voltage sensor to detect neural activity. Sci Rep 9, 15878 (2019). doi: 10.1038/s41598-019-52370-8
Resources on Addgene.org
Resources on the Addgene blog
Topics: Viral Vectors 101





Leave a Comment