This article was written by Alyssa Cecchetelli and Andrew Hempstead.
What do smell, taste and sight have in common, besides being one of the five senses? GPCRs or G-protein-coupled receptors (GPCRs)! Aside from these three senses, GPCRs play roles in initiating signaling pathways in inflammation, and neurotransmission. These receptors can be activated by an array of different ligands, including hormones, neurotransmitters, photons or odorants, to elicit downstream signaling cascades.
So let’s break down the name "G-protein coupled receptor" into two parts: the receptor itself and the G-proteins it associates with.
- Receptor: GPCRs are also known as seven-(pass)-transmembrane domain receptors, due to the seven alpha helices that transverse the cellular membrane. The receptor recognizes the appropriate ligand to activate the G proteins. It is estimated that there are approximately 950 different human genes encoding GPCRs (Takeda et al., 2002).
- G proteins: The receptor is coupled with heterotrimeric G proteins on the intracellular side of the cell membrane. Heterotrimeric G proteins are important molecular switches in signal transduction pathways and consist of a trimer of three subunits: alpha (Gα), beta (Gβ), and gamma (Gγ). G proteins are divided into four subtypes, based on their Gα subunit: Gαs, Gαi/o, Gαq/11, and Gα12/13. Gβ and Gγ are closely associated and function as one unit: Gβγ. In general, GPCRs show a propensity to associate with certain subtypes, although most are able to signal through more than one subtype.
How GPCR signaling works
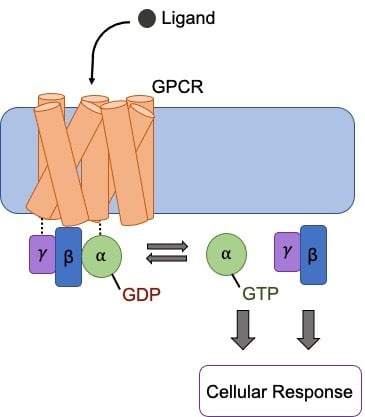
GPCR signaling is initiated when a ligand binds to the extracellular surface of the GPCR. This results in a conformational change in the GPCR causing the activation of the Gα subunit. The activated Gα exchanges bound GDP with GTP, resulting in the disassociation of the Gα subunit from the Gβγ dimer. The Gα and Gβγ subunits then induce or inhibit intracellular signaling cascades as a response to the extracellular stimuli. Ligand dissociation from the GPCR allows for binding of a new inactive heterotrimeric G protein complex and subsequently another round of signaling.
Addgene Tools for Studying GPCR signaling
So with the hundreds of GPCRs out there, how can scientists decipher the signaling pathways from ligand to receptor to G-protein, and finally, to a response? Let’s take a look at a few examples with plasmids deposited at Addgene.
- PRESTO-TANGO: The ligands that activate many GPCRs are still unknown. To identify some of these unknown ligands and their GPCR binding partner, Bryan Roth’s lab created an open source resource called PRESTO-TANGO (parallel receptorome expression and screening via transcriptional output - TANGO) (Kroeze et al., 2015). PRESTO-TANGO uses a modified Tango beta-arrestin recruitment assay to measure receptor activation for more than 300 GPCRs. In this assay, binding of an agonist recruits a beta-arrestin-TEV protease fusion to a receptor-transcription factor fusion. TEV protease cleave the transcription factor from the receptor, which can then activate expression of a luciferase reporter in the nucleus. The Roth lab showed that this platform accurately identified 120 known GPCR targets demonstrating its potential to discover new ligands for unknown GPCRs. You can find the entire PRESTO-TANGO kit at Addgene!
- TRUPATH: Bryan Roth’s lab also assembled a collection of plasmids to elucidate the role of 14 of the 16 non-visual heterotrimeric G proteins that can be activated downstream of GPCRs. This collection, called TRUPATH, allows researchers to identify specific G proteins activated by ligand-bound GPCRs of interest. The TRUPATH plasmid kit consists of 20 plasmids encoding 14 alpha, 2 beta, and 4 gamma subunits of the heterotrimeric G protein complex that can be used to detect G protein activation via bioluminescence resonance energy transfer 2 (BRET2). Check out our blog post on GPCR signaling using TRUPATH for more information on this system!
- GPCR-APEX: Interested in tracking the downstream signaling pathways mediated by GPCRs? The Kruse Lab constructed plasmids that can be used in peroxidase-catalyzed proximity labeling to quantifiably measure GPCR agonist response in vivo (Paek et al., 2017). In this technique, the receptor fused to APEX2 catalyzes the biotinylation of proteins near the GPCR. These proteins are then identified with mass spec. Using this technique termed “GPCR-APEX” the Kruse lab was able track the activation and internalization of two different receptors- angiotensin II type 1 receptor and the β2 adrenoceptor.
GPCRs in chemogenetics
While there’s still a lot to learn about GPCR signaling, researchers in the meantime have developed GPCRs for chemogenetic studies to probe the relationship between neurons and specific behaviors. Chemogenetics uses genetically engineered receptors to interact with small molecules to elicit a response in a certain cell type. Chemogenetics is similar to optogenetics but instead of light, these genetically modified receptors are activated by small molecules to either activate or inhibit neuronal firing. As GPCRs are the largest class of signal transducing receptors in the brain they are ideal candidates for use in chemogenetics.
To create chemogenetics receptors, GPCRs were first mutated via site-directed mutagenesis to bind non natural ligands. These non natural ligands however exhibited off-target effects thus scientists then modified some GPCRs further to respond to specific, pharmacologically-inert, small molecules. These new receptors were termed Designer Receptor Exclusively Activated by Designer Drugs (DREADDs) (Roth, 2016). DREADDs were engineered from members of the human muscarinic receptor family and are activated by small molecule ligands such as clozapine-N-oxide (CNO). DREADDs are ideal for chemogenetics as they are relatively insensitive to their endogenous ligand, acetylcholine, and are only activated upon binding of CNO thus showing no constitutive activity. There are an array of DREADDs that activate different G proteins, work in different neurons and neuroglia, and have different effects on neuron activity (firing or inhibition). AAV preps from DREADD plasmids allow you to easily express a DREADD receptor in a specific neuronal subtype in the brain that can either activate or inhibit neurons. You can find information more about chemogenetics and a concise table of DREADDS and their activity in neurons on Addgene’s chemogenetics guide.
References and resources
References
Kroeze WK, Sassano MF, Huang X-P, Lansu K, McCorvy JD, Giguère PM, Sciaky N, Roth BL (2015) PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol 22:362–369 . https://doi.org/10.1038/nsmb.3014
Paek J, Kalocsay M, Staus DP, Wingler L, Pascolutti R, Paulo JA, Gygi SP, Kruse AC (2017) Multidimensional Tracking of GPCR Signaling via Peroxidase-Catalyzed Proximity Labeling. Cell 169:338-349.e11 . https://doi.org/10.1016/j.cell.2017.03.028
Roth BL (2016) DREADDs for Neuroscientists. Neuron 89:683–694 . https://doi.org/10.1016/j.neuron.2016.01.040
Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S (2002) Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Letters 520:97–101 . https://doi.org/10.1016/s0014-5793(02)02775-8
Additional resources on the Addgene blog
- Learn more about TRUPATH
- Read our quick guide to chemogenetics
Resources on Addgene.org
Topics: Chemogenetics, Neuroscience






Leave a Comment