In 2005, Boyden et al. described the first light-activated tool for controlling neuronal activity, channelrhodopsin-2 (ChR2), a blue light-activated cation channel, from the archaebacteria Chlamydomonas reinhardtii. When exposed to blue light, this channel activates neuronal activity by opening and allowing positively charged ions to rapidly pass through the cell membrane and initiate an action potential - a very useful tool for neuroscientists! Since then, scientists have developed many optogenetic tools to control cellular activity with exquisite precision using only light. Optogenetics, or light activated tools, can control everything from electrical activity to gene expression, and even CRISPR based gene editing. But, how do you decide which tools to use, and what do you need to know about them to be successful in your experiments? In this post, we will discuss some broad classes of optogenetic tools, and what factors to consider when choosing one.
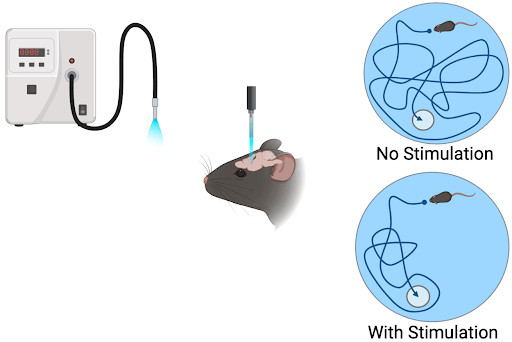
|
| Figure 1: Example of an experimental optogenetic setup. A light source delivers light pulses to the subject/cells expressing the optogenetic tool causing a change in cellular activity or gene expression. Experimental output is measured and quantified. Created with BioRender.com. |
Optogenetic tools
Types of control
We will discuss three types of tools: light-gated ion channels, optogenetic G-protein coupled receptors (OptoXRs), and photoswitches. Light-gated ion channels directly open and close when exposed to light and change the membrane potential. OptoXRs are modified GPCRs. When exposed to light, they activate downstream signaling pathways that can change both the membrane potential and a host of other cellular functions. Photoswitches are engineered molecules that induce conformational changes with light stimulation to either activate or inhibit a specific receptor or pathway.
Light-gated ion channels
Light activated ion channels (rhodopsins) either excite or inhibit neuronal activity upon exposure to light within a specific range of wavelengths. Excitatory channels allow positively charged ions such as Na+, K+, and Ca++ to passively enter the cell with the concentration gradient to depolarize it (Nagel et al., 2003). Inhibitory opsins are usually light-driven pumps that pass ions (Cl-, H+, Na+) against the concentration gradient and hyperpolarize the cell (Mattis et al., 2012). Bicistronic vectors combine excitation and inhibition by using both excitatory and inhibitory channels activated at different wavelengths of light (Vierock et al., 2021). Step-function opsins are chimeric ion channels that turn on with a light-pulse at one wavelength and stay on until a second pulse of light, with a different wavelength, turns it off, allowing the cell to be excited over a sustained period of time (Gong et al., 2020).
Light-gated ion channels are useful when you want to activate or inhibit specific neurons without needing to consider secondary signaling pathways. There are many different opsin (natural and engineered proteins responsive to light in combination with retinal) variations designed to give researchers flexibility in using optogenetic tools in their experiments. Some tools turn on and off rapidly with light to control precise timing of the perturbation. Other tools are designed to be activated within a narrower band of wavelengths to allow multiple tools, such as biosensors, to be used in tandem with the opsin.
-min.png?width=512&height=304&name=unnamed%20(1)-min.png)
|
| Figure 2: Diagram of excitatory (ChR2) activation by light showing ions flowing with the concentration gradient, and of activation inhibitory (ArCH) proton pump passing ions against the concentration gradient. Created with BioRender.com. |
Opto-G-protein coupled receptors
G-protein coupled receptors (GPCR) regulate cellular function through multiple intra-cellular signaling cascades. Optical methods for controlling GPCR activation have been developed to selectively study individual types of GPCRs with greater spatial and temporal control than is achieved by traditional pharmacologic methods.
A number of strategies have been developed for utilizing light to control GPCR activation and downstream signaling cascades (reviewed in Abreu & Levitz, 2020), including light-sensitive chimeric GPCRS (OptoXRs) (Tichy et al., 2019), photoswitchable tethered ligands (PTL), and photoswitchable, orthogonal, remotely-tethered ligands (PORTLs).
OptoXRs are chimeras of the extracellular domain of endogenous light-sensitive receptors, like mammalian rhodopsin, and the intracellular domain of an endogenous GPCR, such as adrenergic receptors. When exposed to light, there is a conformational change in the extracellular domain which activates the native signaling cascade of the intracellular part of the receptor, similar to what the endogenous binding partner would do. This allows precise control over when and where the receptor is activated.
Similarly, PTLs are special, covalently linked molecules (i.e. photoswitches) attached near the binding pocket of an engineered receptor that change shape with light exposure to activate them. PORTLs use a similar concept, except the ligand is attached to a protein binding partner or nanobody that targets the native receptor allowing the endogenous signaling pathway to remain intact and be studied without confounding issues associated with heterologous expression of engineered receptors. OptoXRs are great for studying the broad effects of activating a particular receptor or when a signaling pathway is unknown.
-min.png?width=512&height=409&name=unnamed%20(2)-min.png)
|
| Figure 3: Chimeric OptoXR incorporating rhodopsin undergoes a conformational change when exposed to blue-light and activates downstream secondary signaling pathways. Photoswitchable tethered ligands hold the ligand in close proximity to the receptor, when exposed to light, the ligand engages the binding pocket and activates the receptor. Created with BioRender.com. |
Controlling secondary pathways with photoswitches
OptoXRs are great for activating entire signaling pathways, but what if you’re interested in studying specific components of those signaling pathways? Here’s where photoswitchable intracellular proteins come into play. Photoswitches can be used to activate or suppress secondary signaling pathways or particular targets with light. This is achieved by coupling endogenous signaling molecules with light-activated partners such as light, oxygen, and voltage (LOV) domains, phytochrome B (PhyB), and cryptochrome 2 (CRY2) (Zhang & Cui, 2015). When stimulated with light, the endogenous proteins activate their signaling pathways allowing researchers greater control over when and where to activate the pathway than traditional pharmacology techniques allow. Multiple photoswitches can be used in a single pathway to activate various protein regulators at different points in the pathway. Photoswitches are great for teasing out important branch points along a signaling cascade that may go awry in a disease state.
-min.png?width=512&height=277&name=unnamed%20(3)-min.png)
|
| Figure 4: Example of an intracellular signaling pathway activated by an optogenetic LOV system. In the absence of light the LOV domain blocks the enzyme binding domain preventing downstream signaling pathways. Light exposure removes the block provided by the LOV domain allowing the pathway to be activated. Created with BioRender.com. |
Important considerations for using optogenetic tools
Once you’ve selected the class of tool appropriate for your experiment, you’ll need to find a specific tool. There are many, many options to consider, and the following factors will help you determine which one(s) are candidates for your experiments.
Wavelengths
Most vectors have an optimal wavelength to use but can be activated by a range of wavelengths. While this may give you a little flexibility when activating the optogenetic tool, it can also cause problems when using multiple optogenetics or biosensors in one system. There can be substantial overlap in the activation curves (Marshel et al., 2019) for each tool, and you may have to test and optimize the intensities of light sources used to prevent activation of both tools with each light source used.
Channel and activation kinetics
Some channels turn on and off very rapidly in response to a single light pulse whereas others have slower responses (Gunaydin et al., 2010). This can affect how rapidly you can activate or inhibit a cell in succession and maintain fidelity with the stimulus. For example, if you want to stimulate at a fast frequency without missing an action potential, then a channel with fast kinetics may be required, whereas a channel with slower kinetics can be used for lower frequency stimulation.
GPCR signaling
GPCR signaling is a complicated process involving many downstream signaling mechanisms that are often cell-type specific. If it is something you’re considering for your experiment, note that you cannot extrapolate results from one cell type to another, as they may not use the same pathways. Additionally, as GPCR signaling is amplified in successive steps along a pathway, there can be very different responses to long vs. short stimulation times or frequencies (Zhang & Cui, 2015). You’ll need to test for these differences when designing your experiment.
Finding the right information
When using a previously published system in your own work, you’ll need to know (and understand!) the hardware types, construction, and testing used for delivering your light stimulus. This can make or break your experiments!
First, closely read the publication, including the supplemental data. You’re most likely to find thorough characterization data, like spectral and kinetic activation information, in the supplemental data instead of in the main text.
Then, check out the Deisseroth lab’s Optogenetics Resources and the Boyden lab’s Synthetic Neurobiology Group Resources, both of which have extensive collections of publications and information on hardware, optogenetics tools, viruses, and more.
Conclusion
Optogenetic tools provide a powerful way to study multiple aspects of cellular activity and physiology. However, there are a number of factors that can wreak havoc on your experiments, so it is important to understand the details of how each tool performs and the consequences of off-target activity. Whether you are studying neuronal signaling or cancer pathways there are a number of optogenetic tools now available to help light your path!
Resources and References
Recommended reading
Abreu, N., & Levitz, J. (2020). Optogenetic Techniques for Manipulating and Sensing G Protein-Coupled Receptor Signaling. In D. Niopek (Ed.), Photoswitching Proteins (Vol. 2173, pp. 21–51). Springer US. https://doi.org/10.1007/978-1-0716-0755-8_2
Lehtinen, K., Nokia, M. S., & Takala, H. (2022). Red Light Optogenetics in Neuroscience. Frontiers in Cellular Neuroscience, 15, 778900. https://doi.org/10.3389/fncel.2021.778900
Mattis, J., Tye, K. M., Ferenczi, E. A., Ramakrishnan, C., O’Shea, D. J., Prakash, R., Gunaydin, L. A., Hyun, M., Fenno, L. E., Gradinaru, V., Yizhar, O., & Deisseroth, K. (2012). Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nature Methods, 9(2), 159–172. https://doi.org/10.1038/nmeth.1808
References
Abreu, N., & Levitz, J. (2020). Optogenetic Techniques for Manipulating and Sensing G Protein-Coupled Receptor Signaling. In D. Niopek (Ed.), Photoswitching Proteins (Vol. 2173, pp. 21–51). Springer US. https://doi.org/10.1007/978-1-0716-0755-8_2
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G., & Deisseroth, K. (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience, 8(9), 1263–1268. https://doi.org/10.1038/nn1525
Gunaydin, L. A., Yizhar, O., Berndt, A., Sohal, V. S., Deisseroth, K., & Hegemann, P. (2010). Ultrafast optogenetic control. Nature Neuroscience, 13(3), 387–392. https://doi.org/10.1038/nn.2495
Marshel, J. H., Kim, Y. S., Machado, T. A., Quirin, S., Benson, B., Kadmon, J., Raja, C., Chibukhchyan, A., Ramakrishnan, C., Inoue, M., Shane, J. C., McKnight, D. J., Yoshizawa, S., Kato, H. E., Ganguli, S., & Deisseroth, K. (2019). Cortical layer-specific critical dynamics triggering perception. Science, 365(6453), eaaw5202. https://doi.org/10.1126/science.aaw5202
Mattis, J., Tye, K. M., Ferenczi, E. A., Ramakrishnan, C., O’Shea, D. J., Prakash, R., Gunaydin, L. A., Hyun, M., Fenno, L. E., Gradinaru, V., Yizhar, O., & Deisseroth, K. (2012). Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nature Methods, 9(2), 159–172. https://doi.org/10.1038/nmeth.1808
Nagel, G., Szellas, T., Huhn, W., Kateriya, S., Adeishvili, N., Berthold, P., Ollig, D., Hegemann, P., & Bamberg, E. (2003). Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proceedings of the National Academy of Sciences, 100(24), 13940–13945. https://doi.org/10.1073/pnas.1936192100
Tichy, A.-M., Gerrard, E. J., Sexton, P. M., & Janovjak, H. (2019). Light-activated chimeric GPCRs: Limitations and opportunities. Current Opinion in Structural Biology, 57, 196–203. https://doi.org/10.1016/j.sbi.2019.05.006
Vierock, J., Rodriguez-Rozada, S., Dieter, A., Pieper, F., Sims, R., Tenedini, F., Bergs, A. C. F., Bendifallah, I., Zhou, F., Zeitzschel, N., Ahlbeck, J., Augustin, S., Sauter, K., Papagiakoumou, E., Gottschalk, A., Soba, P., Emiliani, V., Engel, A. K., Hegemann, P., & Wiegert, J. S. (2021). BiPOLES is an optogenetic tool developed for bidirectional dual-color control of neurons. Nature Communications, 12(1), 1–20. https://doi.org/10.1038/s41467-021-24759-5
Zhang, K., & Cui, B. (2015). Optogenetic control of intracellular signaling pathways. Trends in Biotechnology, 33(2), 92–100. https://doi.org/10.1016/j.tibtech.2014.11.007
Additional resources on the Addgene blog
-
Chemogenetics vs. Optogenetics: Which Method Should I Choose?
- GPCRs: How Do They Work and How Do We Study Them?
- Optogenetics + CRISPR, Using Light to Control Genome Editing
Additional resources on addgene.org
-
Read our Optogenetics Guide
- Browse Optogenetics Plasmids
Topics: Optogenetics, Viral Vectors, Viral Vectors 101






Leave a Comment