Viruses are simple: a genome packaged in a protein shell (Taylor, 2014). They’re so simple that we can’t even decide if they’re alive or not. Yet these simple, small particles have quite the outsized impact — and not just on the disease front.
Viruses survive by hijacking the machinery of the cells they infect. Once they enter a cell, they treat their viral genome as a “payload” that is delivered to the cell. The cell then uses its own machinery to replicate the viral genome and proteins. This is, for the intrepid researcher looking for molecular biology tools, a very useful trait.
Virus structure
While all viruses have a genome inside a protein shell known as a capsid, some also have a lipid bilayer surrounding the capsid, called an envelope. Viruses without an envelope are referred to as naked or non-enveloped, while viruses with an envelope are called enveloped viruses (Figure 1). Like bacteria, only a tiny fraction of viruses are pathogenic (Balloux & van Dorp, 2017).
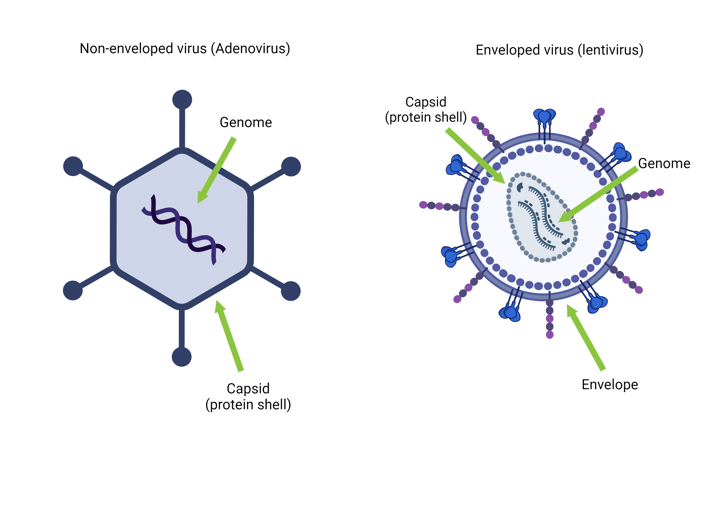
Figure 1: A non-enveloped virus (left) and an enveloped virus (right). Created using biorender.com
If you’re new to talking about viruses, here’s a quick tip on terminology. “Virus” refers to a single type of virus, not the individual viral particles. A single viral particle is called a virion. Finally, viral vectors, which are used in the lab, are distinct from viruses — we’ll get to that in a bit!
Viral genomes and replication
When it comes to replication, viruses tend to march to the beat of their own drum. They have a wide variety of possible genome types that can be either double stranded (ds) or single stranded (ss) (ie, dsDNA, ssDNA, dsRNA, positive sense ssRNA, or negative sense ssRNA) and varying methods of replication. In fact, an entire classification scheme, known as the Baltimore replication classes, has been developed around how they replicate (Figure 2).
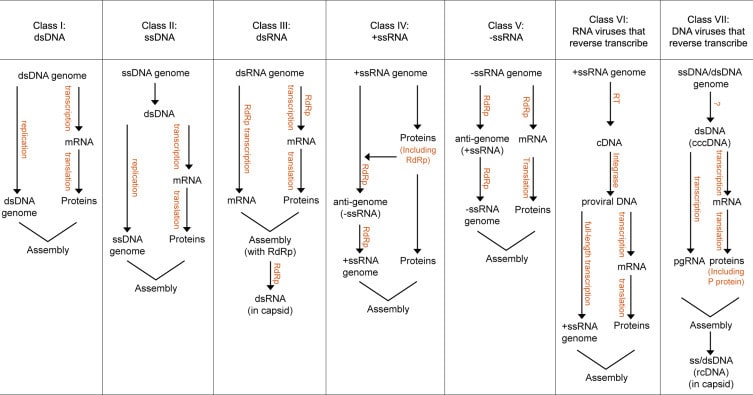
Figure 2: Baltimore replication classes. Image credit: Louten, 2016
Viruses with DNA genomes can either follow central dogma (gDNA to RNA to proteins) or require an intermediary DNA replication step between DNA and mRNA. DNA genome replication can involve an RNA or dsDNA intermediary, depending on the class of virus.
RNA genomes can be transcribed into mRNA, directly translated into proteins, or, in the case of retroviruses, reverse transcribe their gRNA into DNA, which is then transcribed into mRNA before being translated into proteins. RNA viruses replicate their genomes using a variety of RNA or DNA intermediaries, depending on the class of virus.
From one, come many
After a virion infects a cell, the single particle creates many copies of itself, sometimes up to millions of new copies, which are released from the cell like baby spiders from an egg sac. (Okay, some enveloped viruses bud out, instead of lysing, but they do generally still kill the infected cell.) This is in contrast to bacteria and eukaryotes, which use a binary replication process whereby one cell copies itself and “splits” into two cells.
Viral vectors
Viruses are quite appealing as molecular biology tools… if one isn’t too bothered by the risk of infection, that is. Thankfully, through the use of engineering controls — changes made to the virus that eliminate or reduce the risk of infection — researchers were able to create far safer viral vectors that can be used in BSL-1 or BSL-2 labs.
Viral vectors are viruses that have been genetically modified to either limit or completely eliminate their replicative ability. The remaining particle, which retains the protein coat (and envelope), has a “gutted” genome with genes central to the replication process removed. This means the vector can deliver a genetic payload to a cell but cannot create new particles. As a bonus, this increases the carrying capacity of the viral vector, increasing both safety and usefulness of the tool. In fact, most gutted genomes have as much genome removed as possible, just to increase the viral vector’s carrying capacity.
In order to allow the viral vectors to reproduce when experimentally necessary, genes for the required proteins are packaged in separate plasmids and expressed via cells cultured with the vectors. This allows the viral vectors to reproduce only in experimentally controlled conditions, making them far safer to work with than virions.
The four types of viral vectors (which many researchers will informally refer to as viruses) are named after the viruses they’re derived from: AAVs, lentivirus, gamma retrovirus, and adenovirus. And now that you know what viral vectors are, it’s time to dive into how they’re used! We have a growing number of Viral Vectors 101 posts about each type, so be sure to check our blog, or our Viral Vectors 101 eBook, out if you want to learn more.
References and resources
References
Balloux, F., & van Dorp, L. (2017). Q&A: What are pathogens, and what have they done to and for us? BMC Biology, 15, 91. https://doi.org/10.1186/s12915-017-0433-z. PMID: 29052511.
Louten, J. (2016). Virus Replication. Essential Human Virology, 49. https://doi.org/10.1016/B978-0-12-800947-5.00004-1
Taylor, M. W. (2014). What Is a Virus? In M. W. Taylor (Ed.), Viruses and Man: A History of Interactions (pp. 23–40). Springer International Publishing. https://doi.org/10.1007/978-3-319-07758-1_2
More resources on the Addgene blog
Viral Vectors 101: Viral Applications
Viral Vectors 101: Viruses as Biological Tools
Viral Vectors 101: Viral Vector Elements
More resources from Addgene
Topics: Viral Vectors 101, AAV, Retroviral and Lentiviral Vectors, Adenoviral Vectors







Leave a Comment