Every few months we highlight a subset of the new plasmids and viral preps in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. If you'd ever like to write about a recent plasmid deposit please sign up here.
 Here's what you'll find in this post:
Here's what you'll find in this post:
- Blue-light inducible Cre for use in bacteria
- CRISPR-Casɸ
- mGold, a new yellow fluorescent protein
- New CRISPR plasmids
- New items from the viral service
A fast blue-light inducible Cre for precise control of bacterial gene expression
By Meghan Rego
Recombinases are enzymes that recognize, cut, and alter specific target sequences in DNA resulting in a variety of outcomes including excision or insertion, inversion, translocation or cassette exchange. By utilizing recombinases to manipulate genomes, scientists can control expression of their genes of interest. While recombinases are powerful gene expression tools in and of themselves, their activity can be fine-tuned and precisely controlled through the use of optogenetics. Optogenetic tools use light to induce conformational changes in a protein.
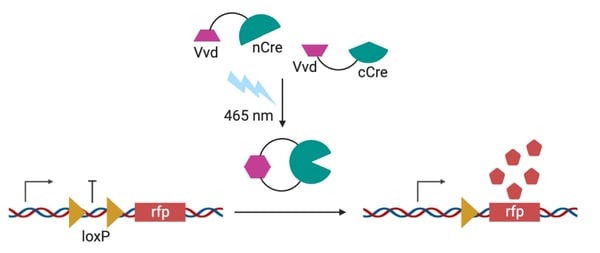 |
| Cells expressing an RFP reporter plasmid for recombinase activity were exposed to blue light. Cre-Vvd showed substantially improved activity as compared to other recombinase/photodimer pairs. Created with BioRender.com |
In their recent ACS Synthetic Biology publication, Mary Dunlop’s lab generated a photosensitive Cre, Opto-Cre-Vvd, by splitting and joining the protein regions with Vivid (Vvd) photodimers. Upon the application of blue light, Vvd dimerizes bringing the C-terminal and N-terminal Cre fragments together and permitting recombinase activity. Opto-Cre-Vvd is precise, demonstrating low sensitivity to ambient light, and versatile as it can be activated by both low and high-intensity blue light. Unlike other split-Cre systems which can take up to 24 hours to cut, Opto-Cre-Vvd is fast and can cut in as little as 2 hours.
Sheets, MB, et al., ACS Synth Biol. 2020. https://pubs.acs.org/doi/10.1021/acssynbio.9b00395
O’Brian, SP, et al., Biotechnol J. 2014. https://pubmed.ncbi.nlm.nih.gov/24390935/
Hypercompact genome editor CRISPR-Casɸ
By Melina Fan
The Doudna Lab adds to the CRISPR-Cas toolbox with their discovery of CRISPR-Casɸ, a hypercompact genome editor from the Biggiephage clade of huge phages. In this article, they investigated three Casɸ orthologs, Casɸ-1, Casɸ-2, and Casɸ-3.
This system is notable for several reasons. Casɸ is only ~70kDa, with a molecular weight roughly half of Cas9 or Cas12a. This is in part due to the fact that it only has a single active site. The RuvC active site in Casɸ can both process crRNA and carry out the crRNA-guided DNA cutting, unlike other well-characterized Cas enzymes, which rely on multiple active sites. In addition, Casɸ has a minimal PAM requirement. For instance, for Casɸ-2 the PAM sequence is 5’-TBN-3’ (where B is G, T, or C). The authors demonstrate that CRISPR-Casɸ is active in vitro as well as in human and plant cells.
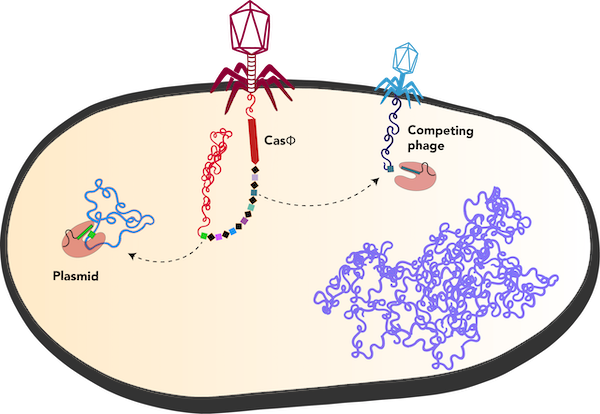 |
| Casɸ from Biggiephage can eliminate competing mobile genetic elements. Image by Basem Al-Shayeb in the Doudna Lab. |
Scientists looking for more compact genome editors due to delivery constraints and scientists seeking a wider range of targetable genomic sequences could benefit from CRISPR-Casɸ.
Pausch et al., Science 2020, https://pubmed.ncbi.nlm.nih.gov/32675376/
mGold, the most photostable yellow fluorescent protein to date
By Justin Ng
High-throughput screening methods, such as fluorescence-activated cell sorting (FACS) helps researchers sift through thousands of cells to identify cells exhibiting properties of interest. However, FACS is unable to spatially and temporally target single cells. Now, Francois St-Pierre’s lab developed a new form of FACS-based cell screening, Single-cell Phenotypic Observation and Tagging with Light (SPOTlight) as a way to isolate individual cells with unique spatiotemporal phenotypes from large heterogeneous cultures.
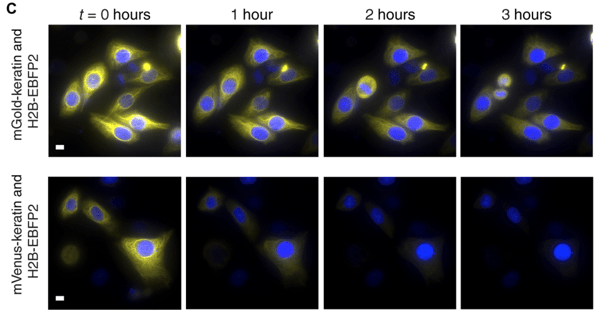 |
| HeLa cells expressed a fusion of keratin with mGold (top) and mVenus (bottom). Cells were imaged every 30 seconds for 3 hours. Image from Lee et al., 2020. |
SPOTlight was used to image over 3 million cell types expressing variants of Yellow Fluorescent Protein (YFP). The team identified a new YFP variant, mGold. mGold was found to be up to 5-fold more photostable and as bright to mVenus, the parent YFP, and is the most photostable YFP to date. As such, mGold is expected to contribute to experiments that require high photostability, such as time-lapse imaging or with high light power detection of molecules.
Lee, J. et al., Science Advances 2020. https://doi.org/10.1126/sciadv.abb7438
The CRISPR corner
Here are a few highlights from recent CRISPR plasmids. To find all of the CRISPR plasmids available from Addgene, head over to our CRISPR Plasmids and Resources page.
- CDDR (CRISPR–Cas9-based Dual-fluorescent DSB Repair) is a double-strand break repair assay for mammalian cells. It's based on the introduction & resolution of two DSBs in a fluorescent reporter.
- Yongsub Kim’s lab engineered prime editors to expand their PAM flexibility.
- A lentiviral CRISPR knockout library that targets human epigenetic genes was deposited by Kivanc Birsoy.
- A new SaCas9 ABE variant (microABEI744) has improved on-target editing efficiency and reduced RNA-off target footprint compared to current N-terminal linked SaCas9 ABE variants. It's also one of the smallest AAV-deliverable ABE.
- Jesse Zalatan’s lab deposited plasmids for a chemically inducible CRISPRa system in yeast is mediated by recruitment of MS2-functionalized gRNAs.
- Reduce the scale of your CRISPR screens by using this lentiviral CRISPR human genome-wide library deposited by Leopold Parts. This library delivers 2 randomly paired guides per construct and allows a reduced scale in screens without any reduction in performance compared to larger genome-wide libraries.
 New from the viral service
New from the viral service
By Jason Nasse
We regularly add new viral aliquots from our plasmid collection to provide ready-to-use viral preps. Here are some of the new viral preps from recent months:
- Expanded serotypes are now available for Gordon Fishell Lab constructs targeting GABA neurons. Serotypes include AAV1, and AAV9.
- pAAV-hSyn-Cre-P2A-dTomato is now available in AAV1 and AAV8, as well as AAV5 and AAVrg.
- AAV-CAG-dLight1.3b from the Lin Tian Lab is now available as a ready to use viral prep in serotype AAV5!
- Looking for a knock-out in your experiments? AAV-flex-taCasp3-TEVp from Nirao Shah's and Jim Well's lab induces cre-dependent apoptosis. Now available as a ready to use viral prep in serotype AAV5.
- The red-shifted channelrhodopsin (C1V1) targeting parvalbumin cortical interneurons is now available as a ready to use viral prep in serotype AAV5.
- Astrocyte specific tight affinity glutamate sensor from the Loren Looger Lab now available in AAV1.
- New Dual and Triple feature INTRSECT viral preps from the Deisseroth Lab now available.
- Coming soon! New AAV serotypes from the Sanford Boye lab with enhanced retinal tropism.
Topics: Hot Plasmids, Plasmids, Other









Leave a Comment