Originally written by Marcy Patrick and Mary Gearing on Mar 12, 2015; updated by Christina Mork, Jul 27, 2020; updated by Susanna Stroik January 24, 2023.
DNA damage drives genome instability and contributes to cancer, premature aging, and other harmful processes. The most dangerous type of DNA lesion results from breakage of both DNA strands - a double-strand break (DSB). DSBs can be caused by intracellular factors such as nucleases and reactive oxygen species, or external forces such as ionizing radiation, chemotherapeutics, and ultraviolet light. In this post, we will describe the mechanism of homology directed repair, one of the repair pathways of DSBs, with a focus on repair for genome modification purposes.

How does homology directed repair fix DNA double-strand breaks?
In eukaryotic cells, repair of DSBs occurs primarily by two pathways: Non-Homologous End-Joining (NHEJ) and Homology Directed Repair (HDR). NHEJ is the canonical homology-independent pathway as it involves the direct ligation of sticky or blunt ends and often introduces small insertions and deletions along the way. HDR uses homologous sequence as a template for repair and is thus the more accurate mechanism for DSB repair. The process is generally error-free provided the DNA template used for repair is identical to the original DNA sequence at the DSB, or it can introduce specific mutations into the damaged DNA if these modifications exist in the template DNA.
There are several unique HDR mechanisms, but all templated-repair pathways have these central steps:
- The 5’ DNA end of the break is resected by nucleases to create a ssDNA 3’ overhang. This will serve as both a substrate for proteins required for strand invasion and a primer for DNA repair synthesis.
- The ssDNA strand then displaces one strand of the homologous DNA donor and can template repair off the other strand; this results in the formation of a DNA structure referred to as the displacement loop (D loop).
- The recombination intermediates can then resolve to complete the DNA repair process
Homology directed repair pathways
HDR mechanisms can be divided into two main camps: non-conservative and conservative. The conservative methods are characterized by use of a homologous donor (e.g., sister chromatid, plasmid, etc.), and consists of three pathways: classical double-strand break repair (DSBR), synthesis-dependent strand-annealing (SDSA), and break-induced repair (BIR). The non-conservative method is single strand annealing (SSA), an error prone mechanism that notably does not require a donor template and thus won’t be covered in detail here.
Classical double-strand break repair (DSBR)
In the classical DSBR pathway, the 3’ ends invade an intact homologous template to serve as a primer for DNA repair synthesis, ultimately leading to the formation of double Holliday junctions (dHJs). dHJs are four-stranded branched structures that form when elongation of the invasive strand “captures” and synthesizes DNA from the second DSB end. The individual HJs are resolved via cleavage in one of two ways - each junction resolution could happen on the crossing strand (horizontally at the purple arrows) or on the non-crossing strand (vertically at the orange arrows). If resolved dissimilarly (e.g. one junction is resolved on the crossing strand and the other on the non-crossing strand), a crossover event will occur; however, if both HJs are resolved in the same manner, this results in a non-crossover event.
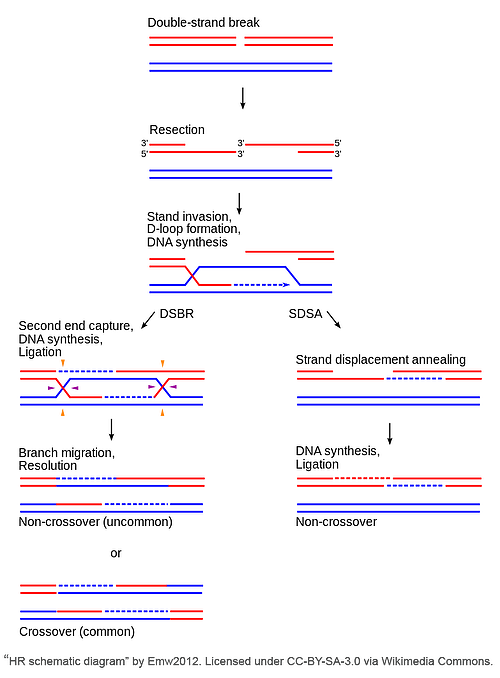
Synthesis-dependent strand-annealing (SDSA) pathway
SDSA is a conservative repair mechanism which exclusively yields non-crossover events. This means all newly synthesized sequences are retained by the original damaged DNA molecule. Unlike in DSBR, following strand invasion and D loop formation, the newly synthesized portion of the invasive strand is displaced from the template and returned to the processed end of the non-invading strand at the other DSB end. The 3’ end of the non-invasive strand is elongated and ligated to fill the gap, thus completing SDSA.
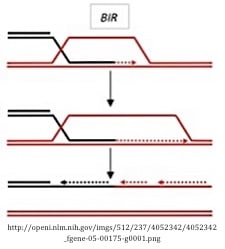
Break-induced repair (BIR) pathway
BIR is characterized by the presence of only one invading strand at a DSB that can be used for repair (a one-ended DSB). This single invasive strand initiates primary synthesis from a template followed by a round of lagging strand synthesis to fill in the resulting ssDNA. A single HJ forms during this process and resolves by cleavage of the crossed strand. While this pathway may not be immediately applicable in DSB-induced genome engineering, it has biological importance for the repair of breaks that lack a second end to enable DSBR or SDSA.
Homology directed repair and genome engineering
CRISPR-Cas9 has revolutionized the genome engineering world and made targeted modifications feasible and even easy. This targeted-break technology coupled with HDR is a powerful site-specific knock-in tool that can be harnessed by researchers for many applications. Your favorite Cas enzyme, a guide RNA to your site of interest, and a donor DNA molecule containing your desired edit are all you need to generate a knock-in.
General considerations for designing a repair template to create mutations
HDR donors can contain insertable sequences or specific mutations, along with homologous DNA to the target sequence to be modified. In general, the insertion sites of the modification should be very close to the DSB, ideally less than 10 bp away if possible. Check out our blog post on Improving HDR efficiency for more details on how "Cut-to-Mutation distance" affects editing efficiency.
One important point to note is that Cas enzymes can continue to cleave DNA if the gRNA/PAM site remains intact. In other words, Cas9 will keep cutting and repairing your target DNA until a mutation is introduced disrupting one or both of these sequences. To prevent re-cutting and the resulting mutations after an HDR event, ensure the gRNA or PAM site is disrupted in the donor.
Choosing a type of donor: plasmid, oligo, double or single stranded?
The size of the intended edit is the biggest determinant when selecting a type of donor. ssDNA templates (referred to as ssODNs (oligodeoxynucleotides)) are commonly used for smaller modifications (~1-50 base pairs). Small edits require as little as 30-50 bases for each homology arm, but keep in mind these numbers may vary based on your locus of interest and experimental system.
Synthesizing ssODNs longer than 200 bases can be difficult, thus dsDNA plasmids are generally preferred for large insertions such as fluorescent proteins or selection cassettes. This class of template should have homology arms between 500-1000 bps. HDR efficiency with circular plasmid templates is generally low; to increase the frequency of edits, researchers have designed self-cleaving plasmids that liberate the targeting region from the vector. Alternatively, PCR-generated or commercially synthesized linear dsDNA templates can be used, however, these are generally more toxic to cells than their plasmid counterparts. Another consideration for dsDNA templates is their ability to randomly integrate elsewhere in the genome, resulting in off-target effects.
A technique that pushes the size limitations of ssODNs is Easi-CRISPR (Quadros et al., 2017). Easi-CRISPR in vitro transcribes RNA encoding your repair template, then uses reverse transcriptase to generate the complementary ssDNA. Easi-CRISPR was shown to increasing editing efficiency from 1-10% with dsDNA to 25-50% with ssODNs in mouse models. Although HDR efficiency varies across loci and experimental systems, ssODN templates generally provide the highest frequency of HDR edits. Check out our blog post on Easi-CRISPR for more details.
References and resources
Want to know more? Check out the additional resources below for more in-depth information on HDR and CRISPR. Our CRISPR 101 series was designed to help explain the basic principles driving CRISPRs and genome editing technology.
Links:
- Zhang Lab’s CRISPR FAQs
- CRISPR forum moderated by the Zhang lab
- CRISPR Challenges: Standardization and HDR
- CrispyCrunch
- Optimizing Donor DNA for Enhanced CRISPR Genome Editing
General Homologous Recombination References:
- Repair of Strand Breaks by Homologous Recombination. Jasin M and Rothstein R. Cold Spring Harb Perspect Biol (2013). PubMed.
- DNA double-strand break repair: how to fix a broken relationship. Pardo B, et al. Cell Mol Life Sci (2009). PubMed.
- Regulation of DNA double-strand break repair pathway choice. Shrivastav M, et al Cell Research (2008). PubMed.
- Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Pfeiffer P, et al. Mutagenesis (2000). PubMed.
- Multiple Pathways of Recombination Induced by Double-Strand Breaks in Saccharomyces cerevisiae. Paques F and Haber JE. Microbiology and Molecular Biology Reviews (1999). PubMed.
- The Road Less Traveled: Strategies to Enhance the Frequency of Homology-directed Repair (HDR) for Increased Efficiency of CRISPR/Cas-mediated Transgenesis (2018). Pubmed.
- The Road Less Traveled: Strategies to Enhance the Frequency of Homology-directed Repair (HDR) for Increased Efficiency of CRISPR/Cas-mediated Transgenesis (2018). Pubmed.
CRISPR References:
- Genome engineering using the CRISPR-Cas9 system. Ran FA, et al. Nature Protocols Nov;8(11):2281-308; (2013). PubMed.
- A Mouse Geneticist's practical guide to CRISPR applications. Singh P, et al. Genetics 114.169771; (2014). PubMed.
- The Use of CRISPR/Cas9, ZFNs, and TALENs in Generating Site-Specific Genome Alterations. Edited by Doudna J and Sontheimer EJ. Methods in Enzymology Vol. 546; (2015). PubMed.
- Optimization of Genome Engineering Approaches with the CRISPR/Cas9 System. Li, K et al. PLoS ONE 9(8); (2014). PubMed.
- Making designer mutants in model organisms. Peng, Y. et al. Development 141; (2014). PubMed.
- Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Richardson CD, et al. Nat Biotech. 34(3); (2016). PubMed.
- Efficient homology-directed repair using single-stranded DNA templates. Jacobi SA, et al. Poster at Keystone Precision Genome Engineering. 2017.
- Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Quadros RM, et al. Genome Biol. 18(1); (2017). PubMed.
Additional Resources on the Addgene Blog
-
Check out our CRISPR Featured Topic Page
-
Learn about Non-Homologous End Joining
-
Learn about Base Editing
-
Read 3 Tips to Improve HDR Efficiency for CRISPR Editing in Human Cells
Resources on Addgene.org
- Browse All CRISPR Plasmids
- Find Validated gRNAs for Your Next Experiment
- Check out our CRISPR Guide
Topics: CRISPR, CRISPR 101







Leave a Comment