We are all oh so familiar with the nuclease Cas9, but how does it stack up to the new Cas enzymes on the block? The Cas family of proteins has many useful genome engineering enzymes, each with their own unique features and quirks. Here, we will review the genome engineering abilities of the lesser-known Cas enzymes while also highlighting their similarities and differences to Cas9.
DNA targeting Cas enzymes
Cas9 – The genome engineering workhorse
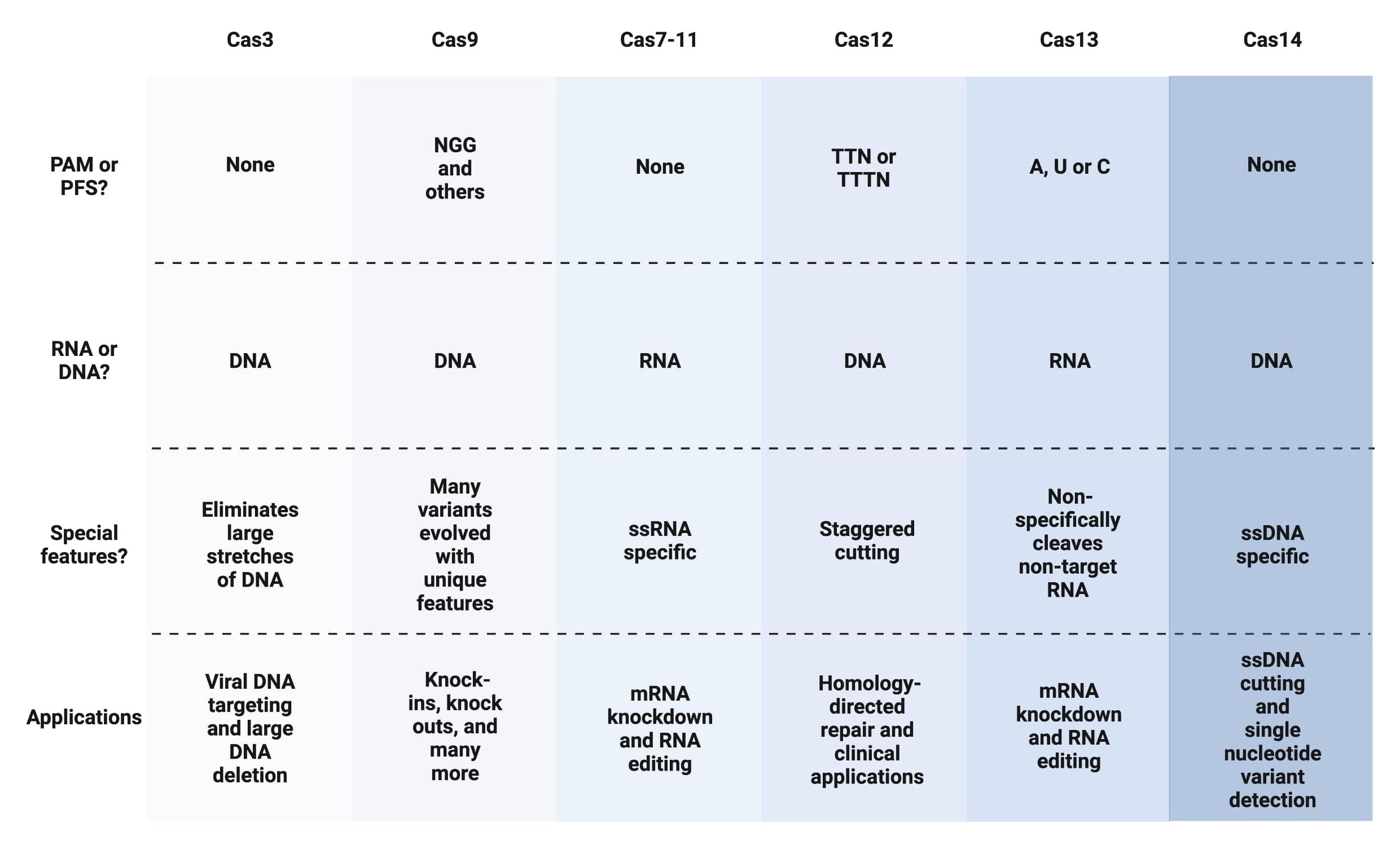
You rarely hear the word ‘CRISPR’ without shortly thereafter hearing ‘Cas9’. The Cas9 nuclease is a genome engineering powerhouse, and for good reason. It is guided by an RNA molecule, the gRNA, to a complementary site in the genome where it will proceed to generate a blunt DNA double strand break. Cas9 has been studied for genome engineering purposes far more than any other Cas family protein and has thus benefitted from countless rounds of optimization. Dozens of variants exist with unique cutting features and evolved PAM sites, making Cas9 an adaptable choice for many genome engineering endeavors. For more information on Cas9 variants, check out our blog post on choosing a Cas9 enzyme.
Fast facts – PAM requirement: NGG and an expanding number of others.
Best for: a wide range of DNA edits.
Cas3 – The shredder
What sets Cas3 apart from Cas9 and almost all other Cas enzymes is its method of DNA cleavage. Instead of cleaving DNA at a defined site to form a clean DNA break, Cas3 locates its target sequence and initiates single stranded DNA degradation. Cas3 DNA digestion can proceed up to multiple kilobases, but only in one direction. Cas3 is thus not helpful for designer genome edits, but it is effective at wiping out large regions of DNA and has anti-viral application potential as a result.
Fast facts – PAM requirement: AAG or TTC
Best for: very large deletions.
Cas12a – The staggered cutter
Originally classified as Cpf1, Cas12’s primary difference from Cas9 is that it generates staggered breaks, also known as sticky ends. Cas12a cleavage results in 5 bp overhangs and is particularly convenient for targeting experiments requiring single strand DNA generation, such as homology directed-repair. Cas12a is also conducive for targeting AT-rich genomic loci as the PAM does not require any G’s or C’s. One curious feature of this Cas is that once it is engaged by its target, it will non-specifically cut ssDNA. This feature is shared by other Cas enzymes outlined below as well.
Cas12a also has significant therapeutic promise due to the fact that it is smaller than the traditional SpCas9 enzyme (about a third of the size) and does not require a tracrRNA (it can self-process pre-crRNA into crRNA). The size and simplicity of Cas12a makes it easier to package for therapeutic delivery by AAV, which has strict capacity limitations. New efforts have recently been made to increase cutting efficiency and diversify PAM sequences of Cas12a to further optimize it for therapeutic and laboratory usage.
Fast facts – PAM requirement: TTN or TTTN.
Best for: homology-directed repair or therapeutic delivery.
Cas14 – Mini, but single strand mighty
Unlike many other Cas enzymes, Cas14 targets ssDNA instead of dsDNA. It cleaves ssDNA with high fidelity - sensitive to even a single mismatch in the target sequence Similar to Cas12, it is also significantly smaller in size than traditional Cas9. Another similarity to Cas12a is that Cas14 also indiscriminately cleaves non-complementary ssDNA when engaged by its target sequence. Why would you want to give it a go? The high specificity of Cas14 makes it a promising detection tool for rare, often hard to detect variants such as single nucleotide polymorphisms.
Fast facts – PAM requirement: none.
Best for: high fidelity ssDNA targeting.
RNA targeting Cas enzymes
Move over Cas9, CRISPR isn’t just for DNA anymore! Cas enzymes have been isolated which target and cleave RNA through the same mechanism as traditional spCas9. If you are looking to modify RNA, Cas9 just won’t cut it – literally.
Cas13
Cas13 is an RNA-guided nuclease, however, unlike Cas9, it targets RNA instead of DNA. This makes it particularly useful in generating transient edits of signaling molecules (RNA) as opposed to permanent edits to the genome. Cas13, similar to Cas14, also indiscriminately cleaves ssRNA when engaged at its RNA target sequence. The RNA-targeting ability of Cas13 makes it perfect for targeting mRNA and useful as a mechanism of transient knockdown of gene expression. This system could be used in lieu of siRNA or shRNA for efficient, multiplexable RNA knockdown. Cas13 has also stirred up interest for its potential as a viral therapeutic that could specifically degrade target viral RNA. For more information on Cas13, check out our blog on RNA editing.
Fast facts – PAM requirement: protospacer flanking sequence – A, U, or C.
Best for: targeting single stranded RNA.
Cas7-11
Cas7-11 is a naturally occurring RNase composed of the fusion of several Cas7 and Cas11 domains. Cas7-11 cuts ssRNA and is capable of pre-crRNA self-processing. Functionally, it is very similar to Cas13 and has been proposed to be useful for the same applications (RNA knockdown, RNA editing, etc.). The main differences between Cas7-11 and Cas13 are 1). non-specific RNA cleavage and 2). cell toxicity. Cas7-11 does not display collateral RNA cleavage and is significantly less toxic to mammalian cells than Cas13 (likely due to the lack of collateral cleavage). You can read about Cas7-11 in our blog post on its development and use.
Fast facts – PAM requirement: protospacer flanking sequence – none.
Best for: targeting single stranded RNA.
|
|
|
Table 1: Fast facts on Cas family proteins and genome editing abilities |
Choosing a Cas
While Cas9 may be the most adaptable Cas currently available, the other Cas proteins should not be overlooked. The unique features and targeting abilities of the other Cas family members make for useful implements in the scientist toolbox. If your genome engineering goals expand beyond generating frameshift mutations or single base pair edits, these enzymes are worth taking a look at. Choosing the right Cas can really put your experiments Nsync.
References and resources
References
Dolan, E., Wang, Hou, Zhonggang, Xiao, Yibei, et. al. Introducing a Spectrum of Long-Range Genomic Deletions in Human Embryonic Stem Cells Using Type I CRISPR-Cas. Molecular Cell, 74(5): 936-950.e5 (2019). 10.1016/j.molcel.2019.03.014
Zetsche, Bernd, HGootenberg, S., Jonathan, Abudayyeh, O., Omar, et. al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163(3): 759-771 (2015). 10.1016/j.cell.2015.09.038
Harrington, B., Lucas, Burstein, David, Chen, S., Janice, et. al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science, 362(6416): 839-842 (2018). 10.1126/science.aav4294
Ozcan, Ahsen, Krajeski, Rohan, Ioannidi, Eleonara, et al., Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature, 597, 720-725 (2021). 10.1038/s41586-021-03886-5
Additional resources on the Addgene Blog
- CRISPR 101: Which Cas9 do I choose for my CRISPR experiment?
- The PAM requirement and expanding CRISPR beyond spCas9
- Cas proteins
More resources on the Addgene Website
Topics: CRISPR 101





Leave a Comment