Note: Cpf1 is also called Cas12a.
There’s a new development for CRISPR-Cpf1 genome editing! A recent paper from Feng Zhang's lab describes how to use Cpf1 for multiplex genome editing. For a few reasons, Cpf1 is a simplified system for editing multiple targets compared to Cas9. Read on to learn more about Cpf1 multiplexing. For an in-depth review of Cpf1, check out this blog post or see Addgene's CRISPR guide page for a review of Cas9. For a brief comparison of Cpf1 vs. Cas9, see the table below.
Table 1. Comparing the Cas9 and Cpf1 CRISPR Nucleases
| Cas9 | Cpf1 | |
| Components | crRNA tracrRNA Cas9 |
crRNA |
|
Endonuclease size |
spCas9: ~4 kb |
~3.9kb |
| crRNA/gRNA length | gRNA: ~100 nt | crNA: ~42 nt |
| Requires tracrRNA? | Yes | No |
| dsDNA cleavage | Blunt end | 5' overhang |
| PAM sequence | NGG | TTTV |
| PAM site preserved? | Usually Destroyed | Yes, Cpf1 cleaves 5' of the protospacer |
Multiplexing CRISPR-Cas9 options before Cpf1 crRNA array
Prior to this new Cpf1 multiplexing method, other multiplex CRISPR gene editing methods relied solely on Cas9. Overall, these approaches have two main drawbacks:
1) Most rely on transfection of more than one vector to express the gRNAs and Cas9. Co-transfections can lead to variable expression levels due to differences in copy number. And there are the usual transfection drawbacks: transient expression and needing to work with a transfectable cell line.
2) They require larger expression vectors which are often more difficult to transfect or package into viral vectors. Cas9 multiplexing vectors are larger because they require regulatory sequences to allow for expression of multiple gRNAs (i.e. Csy4 cleavage sequence, tRNAs, multiple individual promoters). spCas9 and its gRNAs are also larger than their Cpf1 counterparts.
Table 2. Cas9 Multiplexing Options
| Multiplexing Method | Delivery Method | Advantages | Disadvantages | References |
| Yamamoto Lab Golden Gate Assembly |
transfection |
One vector expresses Cas9 and up to 7 gRNAs |
Each gRNA requires its own promoter |
|
|
Multiplexed Lentiviral Expression Cassettes |
lentiviral transduction |
One vector expresses Cas9, eGFP, and up to 4 gRNAs | Each gRNA requires its own promoter | Kabadi et al |
| Csy4-Cleavable Cassettes | transfection | gRNAs expressed from a polycistronic transcript | Requires co-transfection of Cas9 | Nissim et al Tsai et al |
| PTG Cassettes | transfection | gRNAs expressed from a polycistronic transcript | Requires co-transfection of Cas9 | Xie et al |
Multiplexing crRNA expression with Cpf1
The key advantage of multiplexing crRNA expression with Cpf1 is that Cpf1 can process its own pre-crRNA arrays. Zetsche et al demonstrate this by showing that Cpf1 can cleave an array of 4 crRNAs in vitro and when expressed in 293 cells. This allows for a single promoter to drive expression of a crRNA array and doesn’t require the expression of other endonucleases, like Csy4, or the inclusion of processing signals, such at tRNAs, to process the array into functional crRNAs.
Pro-Tip
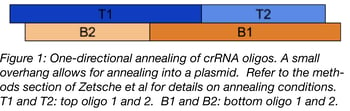
 Cpf1’s ability to process its own pre-crRNA arrays simplifies the crRNA cloning process. For cloning, Zetsche et al used four oligos that consist of direct repeats and crRNA. Similar to a jigsaw puzzle, the oligos were designed with sticky ends that only anneal together in one direction. See the diagram below for an example of how this works. Make sure to order 5’ phosphorylated oligos or treat with T4 PNK.
Cpf1’s ability to process its own pre-crRNA arrays simplifies the crRNA cloning process. For cloning, Zetsche et al used four oligos that consist of direct repeats and crRNA. Similar to a jigsaw puzzle, the oligos were designed with sticky ends that only anneal together in one direction. See the diagram below for an example of how this works. Make sure to order 5’ phosphorylated oligos or treat with T4 PNK.
Delivery options for Cpf1 multiplexing: Transfection, lentivirus, and AAV
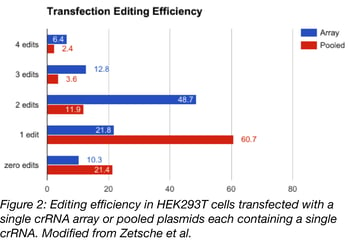 Cpf1’s ability to process its own pre-crRNA arrays eliminates the need to include multiple promoters to drive crRNA expression and sequences that allow for array processing, i.e. tRNAs or Csy4 cleavage sites. This eliminates bulk from crRNA expression plasmids resulting in smaller, more streamlined vectors which are easier to use across multiple expression platforms: transfection, lentiviral transduction, and AAV transduction. In most of Zetsche et al’s experiments, a single vector co-expressing Cpf1 and a crRNA array were used. With transfection, 6.4% of HEK293T cells had edits at 4 of 4 targets when an array of 4 crRNAs, Cpf1, and a GFP tag were transfected on a single plasmid. This is versus 2.4% of cells transfected with a pool of plasmids that contained one crRNA per plasmid plus a Cpf1 expression vector. See the graph in figure 2 for a comparison of editing frequency resulting from transfection of single plasmids vs pooled plasmids; for multiple edits, single plasmids are generally more efficient.
Cpf1’s ability to process its own pre-crRNA arrays eliminates the need to include multiple promoters to drive crRNA expression and sequences that allow for array processing, i.e. tRNAs or Csy4 cleavage sites. This eliminates bulk from crRNA expression plasmids resulting in smaller, more streamlined vectors which are easier to use across multiple expression platforms: transfection, lentiviral transduction, and AAV transduction. In most of Zetsche et al’s experiments, a single vector co-expressing Cpf1 and a crRNA array were used. With transfection, 6.4% of HEK293T cells had edits at 4 of 4 targets when an array of 4 crRNAs, Cpf1, and a GFP tag were transfected on a single plasmid. This is versus 2.4% of cells transfected with a pool of plasmids that contained one crRNA per plasmid plus a Cpf1 expression vector. See the graph in figure 2 for a comparison of editing frequency resulting from transfection of single plasmids vs pooled plasmids; for multiple edits, single plasmids are generally more efficient.
Pro-Tip
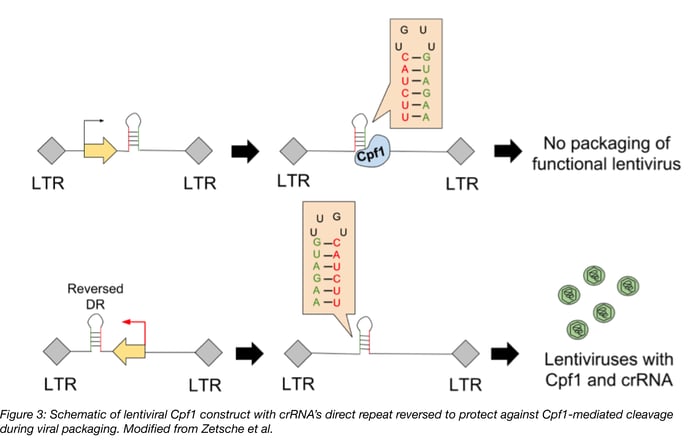
 Make sure to reverse the orientation of the crRNA’s direct repeats when expressing the Cpf1 multiplex system from a lentiviral vector. Lentiviruses carry a (+) strand RNA copy of the DNA sequence, which is a suitable substrate for Cpf1. This means Cpf1 can bind and cleave the lentiviral RNA, preventing packaging of the virus. Reversing the orientation of the direct repeat protects the (+) stranded lentivirus RNA from Cpf1-mediated cleavage. This reversing would also prevent processing of the crRNA array when expressed in cells, but notice how Zetsche et al reverses the orientation of the promoter driving expression of the crRNA array (bolded red arrow). This undoes the original reversing of the direct repeats, leading to expression of a crRNA array that can be processed by Cpf1.
Make sure to reverse the orientation of the crRNA’s direct repeats when expressing the Cpf1 multiplex system from a lentiviral vector. Lentiviruses carry a (+) strand RNA copy of the DNA sequence, which is a suitable substrate for Cpf1. This means Cpf1 can bind and cleave the lentiviral RNA, preventing packaging of the virus. Reversing the orientation of the direct repeat protects the (+) stranded lentivirus RNA from Cpf1-mediated cleavage. This reversing would also prevent processing of the crRNA array when expressed in cells, but notice how Zetsche et al reverses the orientation of the promoter driving expression of the crRNA array (bolded red arrow). This undoes the original reversing of the direct repeats, leading to expression of a crRNA array that can be processed by Cpf1.
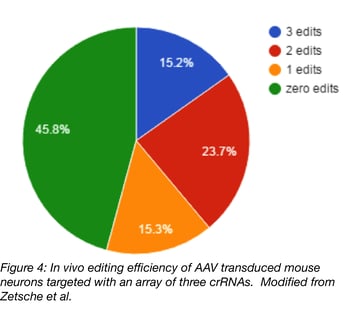
Lastly, an AAV vector was used to multiplex edit 3-4 genes in a primary culture of mouse neurons and in vivo. For these experiments, cells were infected with a 1:1 ratio of two AAVs, one expressing Cpf1 and the other a crRNA array plus a GFP tag. Four weeks post-infection, there was strong expression of Cpf1 and GFP in the targeted brain region and ~75% of neurons were co-transduced with Cpf1 and GFP. Of these neurons, ~15% had indels at all 3 targeted loci. See the chart below for a summary of multiplexing efficiency in vivo.
 Multiplex gene editing with CRISPR-Cpf1 is one of the latest developments in CRISPR technologies. It’s a simple and effective method with multiple applications (in vitro, in vivo, transfection, viral transduction) for simultaneous multi-gene editing with a single crRNA array.
Multiplex gene editing with CRISPR-Cpf1 is one of the latest developments in CRISPR technologies. It’s a simple and effective method with multiple applications (in vitro, in vivo, transfection, viral transduction) for simultaneous multi-gene editing with a single crRNA array.
Are you interested in using Cpf1 multiplexing in your research? Check out the Cpf1 multiplexing plasmids available from Addgene. If you have any questions or thoughts about Cpf1 multiplexing, leave them in the comments below.
References
1. Zetsche et al. "Multiplex Gene Editing by CRISPR–Cpf1 Asing a Single crRNA Array." Nature Biotechnology 35.1 (2016): 31-34. Pubmed PMID: 27918548.
2. Zetsche, B., Gootenberg, J., Abudayyeh, O., Slaymaker, I., Makarova, K., Essletzbichler, P., Zhang, F. (2015). Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell,163(3), 759-771. PubMed PMID: 26422227. PubMed Central PMCID: 4638220.
Additional Resources on the Addgene Blog
- Cpf1: A New Tool for CRISPR Genome Editing
- Which Cas9 Do I Choose for My CRISPR Experiment
- A Match Made in Heaven: CRISPR/Cas9 and AAV
Resources on Addgene.org
- Browse Our CRISPR Multiplexing Resources
- Find CRISPR Plasmids for Your Research
- Catch up on Your CRISPR Background with Our Guide Pages
Topics: CRISPR, Cas Proteins






Leave a Comment