We’ve all heard “Get that tube on ice!!” and “I hope it isn’t degraded” when scientists talk about their precious RNA samples. RNA is inherently less stable than most macromolecules used in scientific research such as DNA or protein. It comes as no surprise then that stability of the RNA used in CRISPR-Cas9 experiments – the “gRNA” – suffers from the same issues as your experimental RNA. To solve this problem, gRNAs can be chemically modified to resist degradation while still effectively serving their function of guiding Cas9 to generate a targeted break. These advances in gRNA technology have enhanced targeting efficacy as well as the unnecessary stress of worrying about the status of your RNA. gRNAs modifications are even going beyond stability, adding color and even on-off switches.
Stabilizing gRNA modifications
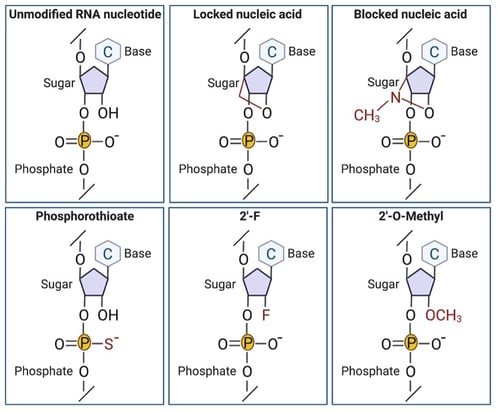
Sugar phosphate backbone modifications
The recognition sequence for CRISPR-Cas9 targeting is led by a ~20 bp gRNA which is part of the larger CRISPR RNA (crRNA). In addition to the 20 bp guide sequence, the crRNA also contains a ~20 bp repeat region on its 3’ end. This repeat region associates with the trans-activating crRNA (tracrRNA) which interacts with the Cas9 protein. These RNA molecules are vulnerable to attack by both cytoplasmic and nuclear nucleases, as they will be recognized as foreign RNA. To protect the RNA from degradation, several simple methods have been employed which modify the sugar or phosphate molecules of the RNA backbone to increase stability.
Nature has evolved ways to prevent degradation of important cellular RNA molecules with post translational modifications. One of the most common of these is 2’-O-methylation, where a methyl group is added to the 2’ hydroxyl group of the ribose to protect it from nucleolytic attack. Another stabilizing sugar modification, found at the same site, is 2’-fluoro (2’-F), which substitutes a fluorine for the hydroxyl group at the 2’ position. Moving one position over in the sugar ring, a popular stabilizing modification is 3’ phosphorothioate linkage, where a sulfur replaces a nonbridging phosphate oxygen. Other useful backbone modifications to consider include amide linkages, unlocked nucleic acids, and a constrained ethyl.
Can’t decide which RNA backbone modification is best? Studies suggest coupling several of them together, for example phosphorothioates with 2’-O-methyl modifications, proved more stability than just one modification alone (Hendel, et al., Nat Biotechnol; Rozners, JACS). These modifications can be done in-lab using commercially available kits or ordered as synthesis modifications when purchasing your gRNA.
Pro tip: modifications are typically incorporated at the termini of the crRNA molecule (usually 1-3 of the first 3 bps are modified) for optimal stability and efficacy.
Base modifications
What is more stable than RNA? DNA! One way to make your gRNA more efficient on a budget is to simply swap out several of the ribonucleotides for deoxynucleotides. A partially DNA gRNA is surprisingly well-tolerated by the Cas9 protein and can increase on-target efficiency. In the same vein, locked nucleic acids or bridged nucleic acids can also be introduced in the gRNA. These modifications result in conformationally restricted RNA which improves mismatch discrimination and is more nuclease-resistant, resulting in less off-target events. While both have similar advantages, bridged nucleic acids with N-methyl substitutions are slightly more efficient and less toxic to mammalian cells (Allen, et al., Frontiers in Genome Editing).
Some of the RNA stabilizing modifications listed above were developed before CRISPR-Cas9, for use in applications like siRNA and antisense oligonucleotides. Some of these synthetic modifications, specifically ones which do not contain a 5’ triphosphate in the gRNA, reduce the innate immune response associated with gRNA introduction (Allen, et al., Frontiers in Genome Editing). This is particularly important in experiments in which viability is being compromised by introduction of foreign RNAs or immunity itself is being studied.
 |
|
Chemical RNA modifications to improve stability.
|
On-off switches for gRNAs
A major drawback of CRISPR-Cas9 technology is that most expression systems are constantly “on”. There is no mechanism to turn cutting off or on at a specific time. Recent advances in gRNA modification have overcome these woes with photoactivatable and photocleavable gRNAs. Photocleavable guides are generated by introduction of a single photocleavable 2-nitrobenzyl linker in the 20-bp targeting region of the gRNA. These gRNAs can be cleaved and rendered useless after less than a minute of exposure to the appropriate light. The photoactivatable counterpart to this system utilizes a gRNA which is photocaged and unable to engage a target sequence. Upon a brief light exposure (several seconds) the gRNA is released and the CRISPR-Cas9 system can initiate targeted editing events. Both technologies utilize chemical modifications to the RNA to control the entire editing system with a simple LED light.
Lighting up your gRNA with fluorescent dyes
In a scientific world where it often seems like everything glows, why not spread the fluorescence to your gRNAs? Dye labeling the RNA in your targeting experiments allows you to visualize transfection efficiency and even track localization in cellular experiments. There are many suitable dyes to choose from and you can opt to do-it-yourself with a kit or purchase it commercially synthesized with a label. Not enough options for you? You can choose to dye label either the crRNA or the tracrRNA. Of course, in a single guide system (crRNA and tracrRNA as a single molecule) you will not have that option, but you can still choose what dye to use
What to do once you’ve chosen a gRNA and modification(s)
Congratulations! You’ve picked your perfect guide; now check out our blog posts on choosing the best Cas9 enzyme to pair with your guide and our available protocols to get your CRISPR experiment moving. Happy editing!
References and Resources
References
Hendel, A., Bak, R., Clark, J. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33, 985–989 (2015). https://doi.org/10.1038/nbt.3290
Rozners, E. Chemical Modifications of CRISPR RNAs to improve Gene-Editing Activity and Specificity. JACS 144, 12584-12594 (2022). https://doi/pdf/10.1021/jacs.2c02633
Allen, D., Rosenberg, M., Hendel, A. Using Synthetically Engineered Guide RNAs to Enhance CRISPR Genome Editing Systems in Mammalian Cells. Frontiers in Genome Editing, 28 (2021). https://doi.org/10.3389/fgeed.2020.617910
Zou, R., Liu, Y., Wu, B., et al. Cas9 deactivation with photocleavable gRNAs. Molecular Cell 81, 7, 1553-1565 (2021). https://doi.org/10.1016/j.molcel.2021.02.007
Moroz-Omori, E., Satyapertiwi, D., Ramel, MC., et al. Photoswitchable gRNAs for Spatiotemporally Controlled CRSIPR-Cas-based Genomic Regulation. ACS 6, 5, 695-703 (2020). https://pubs.acs.org/doi/10.1021/acscentsci.9b01093
More resources on addgene.org
- View Addgene's collection of empty gRNA vectors
- View Addgene's CRISPR resources
- Request ready-to-use viral preps of select CRISPR lentiviral plasmids
Additional resources on the Addgene Blog
Topics: CRISPR gRNAs, CRISPR Protocols and Tips





Leave a Comment