ITRs and serotypes
The names of AAVs contain two numerical designations, separated by a slash, that indicate the ITR and serotype, respectively. For example, AAV2/8 has a type 2 ITR and a type 8 serotype. Almost all vectors contain type 2 ITRs, which comes from the AAV2 serotype. Serotype refers to the type of capsid (outer protein shell) used to package an rAAV vector; these capsids allow the virus to infect (or not infect) different cell types with some specificity. There are a lot of AAV serotypes, many of which are available in rAAVs!
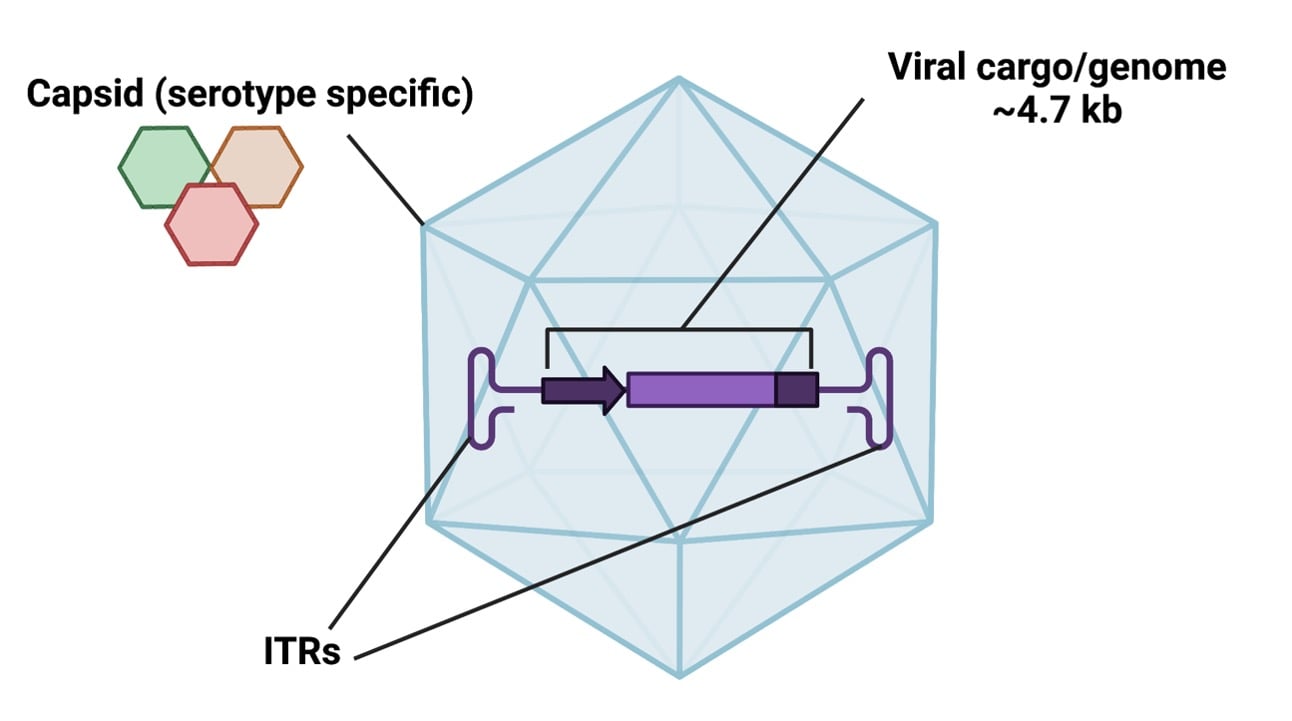
Figure 1: Diagram of AAV components |
Cargo capacity
Empty rAAV has a cargo capacity of ~ 4.7 kb, meaning that your gene of interest (or genes) need to be 4.7 kb or less to fit in a single rAAV. For those interested in heftier genes, you can coinfect with multiple viruses to express genes or gene sets over 4.7 kb. Co-infection rates for AAVs are generally high enough for this approach to work, although it’s less effective in vivo than in vitro.
On the other hand, if you’re interested in expressing multiple small genes, you can design single rAAV vectors with multiple genes packaged in them and use elements such as IRES or 2A to co-express them from one promoter.
Specificity
The ability of AAV to drive gene expression in specific cells and/or tissues is one of its biggest advantages. Specificity can be achieved using specialized promoters, Flp or Cre dependent gene switches, and serotype selection. Promoters and serotype control, respectively, cell-specific gene expression and viral infection. Cre or Flp dependent switches control gene expression via switches placed in your cell type of interest. Careful combination of promoters, gene switches, and/or serotype can also be used to ensure specificity; this is particularly helpful when wanting to target specific neuron subtypes, since AAVs tend to be very neuro-permissive (Issa, 2023).
Time to expression
AAVs have to infect cells, hijack host cell machinery, and synthesize a second strand of DNA before expression of their cargo can begin, which all takes time. Though the time delay until expression is highly dependent on the serotype and target tissue, two weeks is the standard ballpark estimation. Once expressed, viral cargo is usually quite stable and in some mammalian tissues has shown expression for several years (Wonjo et al., 2013). Remember that AAVs do not integrate into the host genome and therefore the transduced gene is episomal and does not divide and replicate in the same way the host DNA does. Because of this, fast dividing tissues will lose expression quicker than terminally differentiated, non-dividing cells.
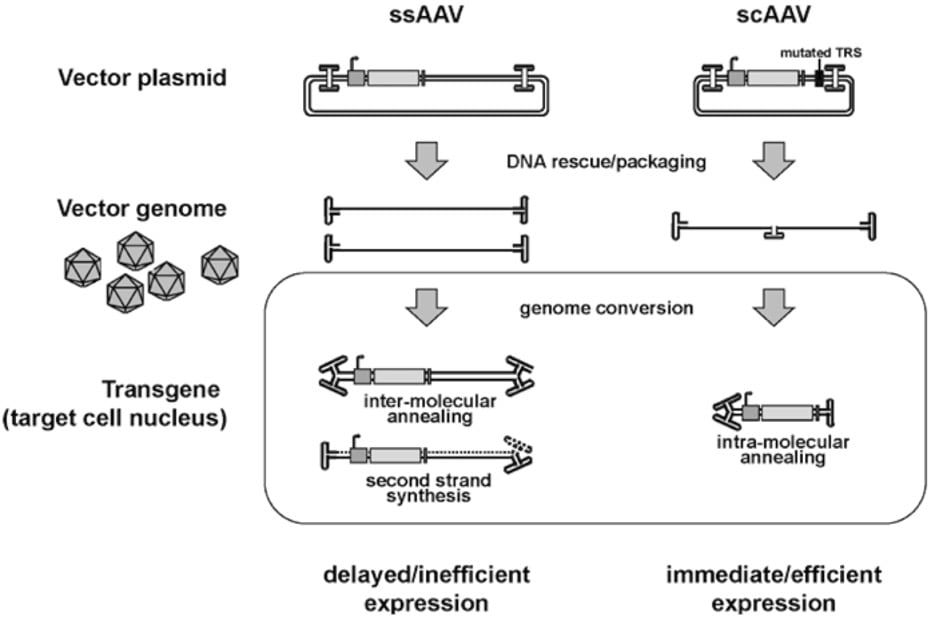 |
Figure 2: DNA rescue and transduction of a conventional single-stranded AAV (ssAAV) and a self-complementary AAV (scAAV) vector. Full-length ssAAV vector genome of both polarities are rescued from the vector plasmid and individually packaged into the AAV capsids. As a genome conversion in the transduced cell nucleus, the single-to-double stranded conversion of the DNA goes through the inter-molecular annealing or second strand synthesis. In contrast, a scAAV vector with half the size of the ssAAV genome has a mutation in the terminal resolution site (TRS) to form a vector genome with wild-type ITRs at the both ends and mutated ITR at the center of symmetry. After uncoating in the target cell nucleus, this DNA structure can readily fold into transcriptionally active double-stranded form through intra-molecular annealing. Figure and legend from Okada, 2013.
scAAV
If your gene of interest is truly small (<2.5Kb) and you need expression in a hurry, self-complementary AAV (scAAV) may be an option (McCarty et al., 2001). This AAV is designed so that the single stranded DNA folds back on itself to become dsDNA in the nucleus and is a good option if you need rapid transgene expression, since you don’t have to wait for the host to replicate the DNA before expression.
Now that you’ve got the basics of AAVs down, you’re ready to take the next step – selecting your AAV and AAV components (serotype and promoter) for use in your experiment! As a starting point, we recommend checking the literature and our AAV data hub to see if others have used relevant viruses for your system.
This post was contributed by guest blogger Didem Goz Ayturk with edits and updates from Addgenies Karen Guerin and Susanna Stroik.
 Didem Goz Ayturk is a Postdoctoral Fellow in Connie Cepko’s Lab investigating neuronal circuits of the retina using viral tools.
Didem Goz Ayturk is a Postdoctoral Fellow in Connie Cepko’s Lab investigating neuronal circuits of the retina using viral tools.
References and Resources
References
Pillay, S., et al. "An essential receptor for adeno-associated virus infection."Nature 4:520(7588):108-12 (2016). PubMed PMID: 26814968.
McCarty, D. M., Paul E. Monahan, and Richard J. Samulski. "Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis." Gene therapy 8.16 (2001). PubMed PMID: 11509958.
Vandenberghe, Luk H., et al. "Efficient serotype-dependent release of functional vector into the culture medium during adeno-associated virus manufacturing." Human gene therapy 21.10 (2010): 1251-1257. PubMed PMID: 20649475. PubMed Central PMCID: PMC2957237.
Okada, T. (2013). Efficient AAV Vector Production System: Towards Gene Therapy For Duchenne Muscular Dystrophy. InTech. doi: 10.5772/53023
Veldwijk, Marlon R., et al. "Development and optimization of a real-time quantitative PCR-based method for the titration of AAV-2 vector stocks."Molecular Therapy 6.2 (2002): 272. PubMed PMID: 12349826.
Xiong, Wenjun, and Constance Cepko. "Distinct expression patterns of AAV8 vectors with broadly active promoters from subretinal injections of neonatal mouse eyes at two different ages." Retinal Degenerative Diseases. Springer International Publishing, 2016. 501-507. PubMed PMID: 26427452.
Issa, Shaza S., et al. “Various AAV Serotypes and Their Applications in Gene Therapy: An Overview.” Cells, 12(5), 785 (2023). PubMed PMID: 36899921.
Resources on the Addgene Blog
- New to Virus Work? Check out Our Primer for Beginners
- Read Our Lentivirus FAQ
- Learn about Viral Vector Elements
Resources on Addgene.org
- Read Our AAV Guide
- Find AAV Vectors
- Browse All Viral Vector Resources
Topics: Viral Vectors, Viral Vectors 101, AAV








Leave a Comment