The Retroviridae (commonly called retrovirus) family — of which HIV is a member — may seem like an unlikely candidate to use as a viral vector, but retroviruses have been developed into some of the most widely used tools in molecular biology. “Retroviruses” is an umbrella term which encompasses 1.) the gamma-retroviruses — that have been developed into retroviral vectors and 2.) a subclass of retroviruses called the lentiviruses (like HIV-1) — that have been developed into lentiviral vectors.
Lentiviruses are sometimes referred to as “complex retroviruses ” (Cullen, 1992) because of their more complicated genomes. Here, we will mostly use the gamma-retroviruses as a simple spokesmodel for the whole family; however, you can use Figure 1 to compare the genomes of these two family members.
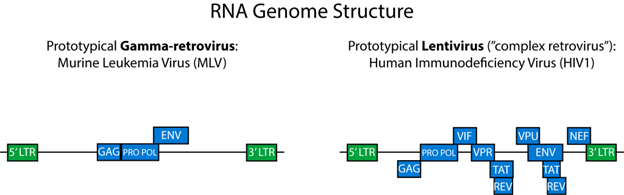 |
| Figure 1: Gamma-retrovirus genome (left) and an HIV lentiviral genome (right.) |
HIV seems an unlikely candidate for use as a tool in the lab because it is, after all, among the more virulent viruses (it attacks the immune system directly). It may seem odder when you consider that out of the 6 x 106 virus-like particles that we come into contact with every day (Prussin et al., 2015), only a small fraction typically cause disease. The gamma-retroviruses have their own unusual sort of virulence as well — namely that they can cause cancer via insertional mutagenesis (which HIV does not do).
Furthermore, molecular biologists working with vectors are, in most cases, interested in transferring genes to mammalian cells (i.e. transferring DNA), yet retroviruses are RNA-containing viruses (Robinson et al., 1965)! You may be beginning to realize that the situation is already complex enough that a deep understanding of retroviral biology is necessary in order to use the vectors correctly. A solid understanding of the structure of a wild-type viral particle — and of the normal, replicative life cycle — can help you understand these seemingly odd choices and lay the foundation for producing and using retroviral vectors effectively.
Retroviral structure
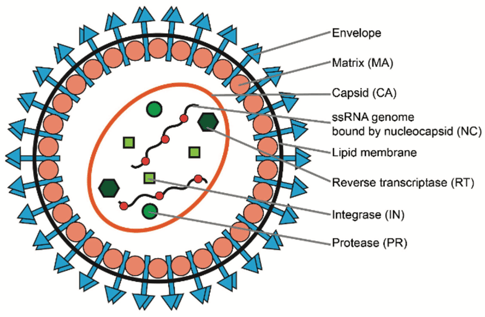
|
| Figure 2: Structure of a retrovirus. Image re-used from Dong & Kantor, 2021 under CC-BY license. |
Figure 2 gives a detailed view of the structure of a retroviral particle, which is essentially the same for viruses and viral vectors. Key to the wild-type virus’ reproductive cycle is the fact that the virus must have its structural proteins expressed from a reverse-transcribed piece of DNA called a “provirus.” This DNA is synthesized inside of what is called the “target cell” (or host cell) by the viral protein reverse transcriptase (RT). In the case of a natural virus, both the protein and the genetic code for reverse transcriptase are packaged (Wu et al., 1997) inside of the capsid . This is useful for viral reproduction, and for causing contagious infection; however this would not be great for a gene therapy vector!
For viral vectors, only the protein for reverse transcriptase (RT) is present in the capsid, as the engineered, recombinant genome is (thankfully) gutted of many essential viral genes. How it is, exactly, that heaps of viral vector particles are made while missing the polymerase gene may seem like a mystery. But it’s simple. The RT protein is made by a separate set of instructions in the same producer cells as the virus. Because it is made at the same time as the other components of the vector particle, some of the RT protein gets packaged inside of the vector particle, along with its RT-deficient genome.
In both cases, the lipid membrane is the only component that is not dependent on viral genes. It is derived from the host/producer cell membrane, and acquired as an envelope during exocytotic budding, as the viral particle emerges.
Retrovirus life cycle
Retroviruses go against the grain of the Central Dogma of Biology (gDNA → mRNA → protein): their roadmap includes RNA → DNA (Cooper & Temin, 1974), as one of its first steps! However, retroviral vectors can still direct the translation of proteins because they integrate this reverse-transcribed DNA into the host cell’s genome and then use cell machinery for transcription and translation. Here, we will discuss several aspects of the viral “life cycle” in order to understand what these vector particles do once they come in contact with target cells (Fig. 3).
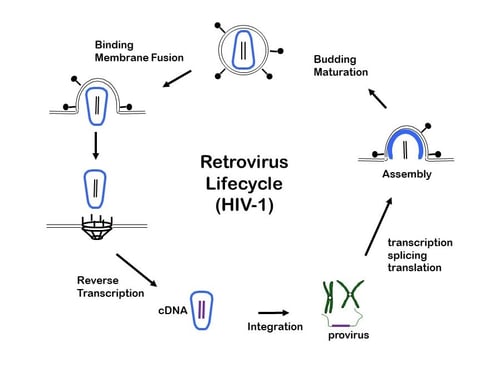
|
| Figure 3: The life cycle of a retrovirus. Image courtesy of Jeremy Luban, MD. |
Figure 3 depicts the viral life cycle. One should ponder this figure often, especially the three steps from the 12 o’clock to the 6 o’clock positions at the left-hand side. These steps are common between natural infection by wild-type viruses and transduction by vectors (which we sometimes still call “infection” as a shorthand, although it is not quite the correct word). Transduction is often thought of as the integration of DNA into the host cell’s genome (shown in step 3), but note also that it is worthwhile to generalize the idea of “transduction” to mean any transfer of material from the virus to the host. In fact, there is a turn of phrase — “pseudotransduction” — to describe the direct protein transfer of RT, which would actually occur in step 2 during uncoating.
After uncoating, reverse transcription follows the release of the two strands of viral RNA and of the viral RT protein (Faras et al., 1973). Interestingly, retroviruses are generally only “pseudodiploid” in that — while they have two dimerized linear ssRNA strands (which can have a different genomic sequence from one another) — by various mechanisms, only one of the two strands’ DNA counterparts from step 2 will be integrated by the viral integrase protein (IN) during step 3 (King et al., 2008) (Hu & Temin, 1990).
At step 3, the double stranded DNA is integrated into the host chromosome, and this is where viral vectors and natural viruses diverge. For the viral vector, its job as a tool is done, and the transgenic genome will simply express from whatever promoters were included in the vector (or are nearby in the host genome). The genome of viral tools are often not virology related at all, and you will instead get expression of the sensor, marker, or whatever other tool you chose to express.
In the case of a natural virus, this new chromosomal locus is considered “proviral DNA” (Rowe et al., 1971) and the full wild-type viral genome can direct the production, packaging, and release of progeny viral particles.
And there you have it, the full “life cycle” of expression of selfish, lazy genes of a non-living viral particle, and how we as dedicated researchers can exploit it.
This blog was written by Addgenie Brian O'Neill, with thanks to Addgenie Rebecca Hutcheson for her assistance with lentiviral replication.
Resources and References
More resources on the Addgene blog
- Viral Vectors 101: Viral Vector Elements
- Viral Vectors 101: Types of Viruses
- Viral Vectors 101: Viral Applications
Resources on addgene.org
References
Cullen, B. R. (1992). Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiological Reviews, 56(3), 375–394. https://doi.org/10.1128/MR.56.3.375-394.1992. PubMed PMID: 1406488. PubMed Central PMCID: PMC372876
Prussin, A. J., Garcia, E. B., & Marr, L. C. (2015). Total Virus and Bacteria Concentrations in Indoor and Outdoor Air. Environmental Science & Technology Letters, 2(4), 84. https://doi.org/10.1021/ACS.ESTLETT.5B00050. PubMed PMID: 26225354. PubMed Central PMCID: PMC4515362
Robinson, W. S., Pitkanen, A., & Rubin, H. (1965). The nucleic acid of the Bryan strain of Rous sarcoma virus: purification of the virus and isolation of the nucleic acid. Proceedings of the National Academy of Sciences of the United States of America, 54(1), 137–144. https://doi.org/10.1073/PNAS.54.1.137. PubMed PMID: 4285422. Pubmed Central PMCID: PMC285811
Wu, X., Liu, H., Xiao, H., Conway, J. A., Hunter, E., & Kappes, J. C. (1997). Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. The EMBO Journal, 16(16), 5113–5122. https://doi.org/10.1093/EMBOJ/16.16.5113. PubMed PMID: 9305652. PubMed Central PMCID: PMC1170145
Cooper, G. M., & Temin, H. M. (1974). Infectious rous sarcoma virus and reticuloendotheliosis virus DNAs. Journal of Virology, 14(5), 1132–1141. https://doi.org/10.1128/JVI.14.5.1132-1141.1974. PubMed PMID: 4372391. PubMed Central PMCID: PMC355630
Faras, A. J., Taylor, J. M., Levinson, W. E., Goodman, H. M., & Bishop, J. M. (1973). RNA-directed DNA polymerase of Rous sarcoma virus: Initiation of synthesis with 70 S viral RNA as template. Journal of Molecular Biology, 79(1), 163–183. https://doi.org/10.1016/0022-2836(73)90277-5. PubMed PMID: 4355596
King, S. R., Duggal, N. K., Ndongmo, C. B., Pacut, C., & Telesnitsky, A. (2008). Pseudodiploid Genome Organization Aids Full-Length Human Immunodeficiency Virus Type 1 DNA Synthesis. Journal of Virology, 82(5), 2376. https://doi.org/10.1128/JVI.02100-07. PubMed PMID: 18094172. PubMed Central PMCID: PMC2258930
Hu, W.-S., & Temin, H. M. (1990). Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proceedings of the National Academy of Sciences, 87(4), 1556–1560. PubMed PMID: 2304918. PubMed Central PMCID: PMC53514
Rowe, W. P., Hartley, J. W., Lander, M. R., Pugh, W. E., & Teich, N. (1971). Noninfectious ARK mouse embryo cell lines in which each cell has the capacity to be activated to produce infectious murine leukemia virus. Virology, 46(3), 866–876. https://doi.org/10.1016/0042-6822(71)90087-0. PubMed PMID: 4332980
Topics: Viral Vectors, Viral Vectors 101, Retroviral and Lentiviral Vectors







Leave a Comment