Two hybrid systems were developed in Saccharomyces cerevisiae in 1989 and are still used extensively to screen for molecular interactions in the cell, including protein-protein, protein-DNA and protein-RNA interactions.
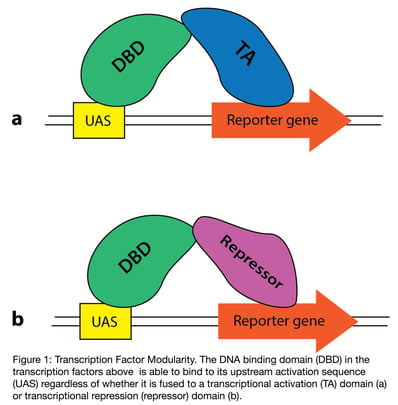 The 1980s saw a flurry of discovery in the field of eukaryotic transcriptional activation and cell biology. During this period, proteins were successfully expressed as fusions that retain their individual activities (1). Researchers also discovered the modular format of some transcriptional activators: that the DNA binding domain (DBD) and transcriptional activation (TA) domains retain their individual activities when separated (2), and that DBD and TAs from different systems could be combined effectively (3). In Figure 1 for instance, the DBD of the fusion protein shown is still able to bind to the upstream activation sequence (UAS) of its target gene, in this case a reporter gene (e.g. luciferase or GFP), regardless of whether it interacts with a transcriptional activator (1a) or a repressor (1b). Scientists took advantage of these discoveries to create a new system for screening protein-protein interactions. In later years, improvements to yeast transformation protocols also helped establish the yeast two-hybrid (Y2H) system as a robust and popular lab tool.
The 1980s saw a flurry of discovery in the field of eukaryotic transcriptional activation and cell biology. During this period, proteins were successfully expressed as fusions that retain their individual activities (1). Researchers also discovered the modular format of some transcriptional activators: that the DNA binding domain (DBD) and transcriptional activation (TA) domains retain their individual activities when separated (2), and that DBD and TAs from different systems could be combined effectively (3). In Figure 1 for instance, the DBD of the fusion protein shown is still able to bind to the upstream activation sequence (UAS) of its target gene, in this case a reporter gene (e.g. luciferase or GFP), regardless of whether it interacts with a transcriptional activator (1a) or a repressor (1b). Scientists took advantage of these discoveries to create a new system for screening protein-protein interactions. In later years, improvements to yeast transformation protocols also helped establish the yeast two-hybrid (Y2H) system as a robust and popular lab tool.
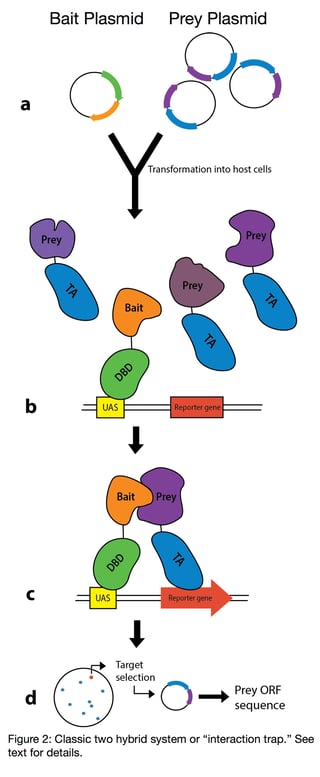
In the classic two hybrid system (also called the interaction trap), genes encoding proteins of interest
(POI’s) are fused to TAs and DBDs (see Figure 2a) (4). These fusions are then co-expressed in a yeast strain that also contains a reporter gene cloned downstream of a promoter containing the target sequence of the DBD. The DBD-POI fusion (commonly referred to as the “bait”, Figure 2b) cannot activate reporter gene expression on its own, but, if the two POI’s interact, this will bring the POI-TA fusion (commonly referred to as the “prey”) in close enough proximity to the promoter of the reporter gene to activate expression (Figure 2c). The reporter gene then allows detection of those cells where binding between the proteins of interest has occurred.
 Although the original Y2H systems utilized the yeast Gal4 activator, bacterial LexA DBD and the lacZ reporter gene (3, 4), there are now many variations, including many that use auxotrophic rescue genes as reporters and some that repress instead of activate transcription. The HIS3 reporter system (e.g. the Gateway-compatible bait Y2H plasmid pEZY202 from Yu-Zhu Zhang’s lab) is an example of the former. In this system, histidine-auxotrophic host strains are used to select for binding-dependent expression of the HIS3 gene, which is required for histidine manufacture and therefore growth on his-deficient media. It is even possible to have Y2H systems that avoid transcriptional reporters altogether; In the split-luciferase system, intein-mediated splicing reconstitutes functional luciferase only when fusions of the two halves of the luciferase protein interact and enable intein splicing (5).
Although the original Y2H systems utilized the yeast Gal4 activator, bacterial LexA DBD and the lacZ reporter gene (3, 4), there are now many variations, including many that use auxotrophic rescue genes as reporters and some that repress instead of activate transcription. The HIS3 reporter system (e.g. the Gateway-compatible bait Y2H plasmid pEZY202 from Yu-Zhu Zhang’s lab) is an example of the former. In this system, histidine-auxotrophic host strains are used to select for binding-dependent expression of the HIS3 gene, which is required for histidine manufacture and therefore growth on his-deficient media. It is even possible to have Y2H systems that avoid transcriptional reporters altogether; In the split-luciferase system, intein-mediated splicing reconstitutes functional luciferase only when fusions of the two halves of the luciferase protein interact and enable intein splicing (5).
Screening with yeast two hybrid systems
While initial studies used single bait and prey protein fusions to demonstrate the utility of the Y2H system, it is more common to use many preys with a single bait in Y2H-based screens. In this way, proteins that interact with a molecule of interest (the ‘bait’) can be ‘fished out’ for further study.
In Y2H screens, a group of prey fusions can either be generated using random genomic fragments from the organism of interest, or a defined cDNA library (e.g. the entire proteome of a particular cell type, or a subset based on a cellular compartment or growth stage). A prey library can either be applied to the bait in a single population (e.g. transforming both fusion plasmids, or mating two single haploid yeast fusion plasmid transformants into a diploid strain for screening), or arrayed on a 96 well plate where each well contains a defined prey fusion. The former allows for screening greater numbers, but the latter eliminates the step of identifying prey proteins after screening.
It is also possible to screen libraries of bait and prey against each other at the same time in order to try and access the entire interactome of a set of proteins at once. However, as Y2H screens are prone to both false positive and false negatives, these experiments must be set up, conducted and analysed with care to get reliable information.
Although two-hybrid systems are a powerful tool for screening molecular interactions, accuracy and reproducibility are common problems when performing these experiments. False positives can occur even in small scale experiments, although these can often be accounted for using appropriate replicates and controls (see Tips below), and by validating leads with other methods. More common are false negatives; up to 75% of known interactions can be missed by single-pass two-hybrid screening (6). See the Tips section for more on how to increase your rate of interactions when screening!
Tips for two-hybrid screening
Minimizing false positives:
- Run replicates of the experiment to reduce the likelihood of indiscriminate reporter gene activation. Including a prey-only control also provides a baseline level of reporter activation.
- Vary the expression levels of bait and prey proteins - overexpression can lead to false interactions. Lowered expression levels will increase the stringency of your binding.
- And most importantly: Independently validate identified binding partners with other techniques!
Minimizing false negatives:
Interactions leading to detectable reporter signals depend on a number of factors, including protein expression and correct folding, post-translational modification, protein degradation, access to the nucleus in eukaryotic screens and fusion configuration. The possibility for problems with one or more of these parameters can lead to a high number of false negatives in Y2H screens.
- Testing your reporter system with a pair of proteins that are known to bind serves as a good positive control to ensure that your setup works. The last thing you want is to run your lovely library screen only to find the system itself is not working because your reporter protein doesn’t function in yeast!
- Many false negatives stem from or are exacerbated by expression of target proteins in heterologous systems (e.g. mammalian proteins in S. cerevisiae).
- While this may be difficult to resolve for a wide range of prey proteins, there are now a number of two-hybrid systems available in model organisms, including bacteria, alternative fungi (C. albicans, pC2HB) (7) and mammalian cells.
- If the problem is that your proteins are not post-translationally modified, co-expression of the enzyme responsible for the modification in the assay host strain can help.
- In order to reduce the chance that the configuration of your fusion proteins physically blocks the binding sites for the protein partner or the UAS/reporter gene, the same bait and prey libraries can be screened using both N- and C-terminal fusions of these proteins. This way both ‘ends’ of your protein are screened for binding.
- Using a variety of expression levels and systems can alleviate false negatives that arise from low or faulty expression of your target proteins. In fact, a good overall strategy to reduce false negatives and produce a more vigourous screen is to use a number of different bait and prey vectors. This has been shown to be as effective as using five independent protein interaction detection methods (6).
- In order to bind DNA and activate a reporter gene in eukaryotic cells, fusion proteins must gain access to the nucleus. To circumvent this requirement for transmembrane proteins, a split ubiquitin system has been devised (8).
References
1. Casadaban M.J., Martinez-Arias A., Shapira S.K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 1983;100:293-308. PubMed PMID: 6312261.
2. Keegan, L., Gill, G., and Ptashne, M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986; 231:699-704. PubMed PMID: 3080805.
3. Brent, R., and Ptashne, M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985; 43:729-736. PubMed PMID: 3907859.
4. Fields, S., Song, O. A novel genetic system to detect protein-protein interactions. Nature. 1989; 340:245-246. PubMed PMID: 2547163.
5. Ozawa, T., Kaihara, A., Sato, M., Tachihara, K., Umezawa, Y.l. Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal Chem. 2001; 73:2516-2521. Pubmed PMID: 11403293.
6. Chen, Y.C., Rajagopala S.V., Stellberger T., Uetz P. Exhaustive Benchmarking of the yeast two-hybrid system. Nature Methods. 2010; 7:667-668. Pubmed PMID: 20805792.
7. Stynen B., Van Dijck P., Tournu H. A CUG codon adapted two-hybrid system for the pathogenic fungus Candida albicans. Nucleic Acids Res. 2010;38(19):e184. Pubmed PMID: 20719741. Pubmed Central PMCID: PMC2965261.
8. Obrdlik, P., El-Bakkoury, M., Hamacher, T., Cappellaro, C., Vilarino, C., Fleischer, C., Ellerbrok, H., Kamuzinzi, R., Ledent, V., Blaudez, D., Sanders, D., Revuelta, J.L., Boles, E., André, D., and Frommer, W.B. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci U S A. 2004; 101(33):12242–12247. Pubmed PMID: 15299147. Pubmed Central PMCID: PMC514463.
Resources At Addgene
- Browse Our Yeast Expression Vectors
- Find Bacterial Two Hybrid Plasmids
- Read our Blog Post About Yeast Vectors
- Read our Blog Post About Tagging Yeast Genes
Topics: Yeast, Molecular Biology Protocols and Tips, Plasmids






Leave a Comment