 There are many, many different types of experiments carried out by scientists every day. Although the designs and outcomes may vary, one thing should be present in every experiment-based investigation of a hypothesis: proper controls!
There are many, many different types of experiments carried out by scientists every day. Although the designs and outcomes may vary, one thing should be present in every experiment-based investigation of a hypothesis: proper controls!
For every experiment, an investigator needs a standard against which the results can be compared; results from an experiment lacking the proper controls are invariably inconclusive and unreliable. Proper controls provide the constant variables that enable the correct interpretation of the effect of the independent variable you are testing. Importantly, they demonstrate the functionality of your experimental system and help identify opportunities for troubleshooting or optimization within your experiment. Read on to learn more about the various controls that can be used for plasmid-based experiments.
 What are control plasmids?
What are control plasmids?
Generally, control plasmids help to ensure that the observed phenomenon in your experiment is specifically associated with the independent variable you are testing and not some other unintended factor. Control plasmids are used to minimize any effects of the non-independent variables.
To illustrate the importance of control plasmids, we will take a step beyond discussion in the abstract and follow an example of a plasmid-based experiment, delineate its requisite control plasmids, and discuss why these control plasmids are critical for the correct design of the experiment.
The Experiment: Knockdown the expression of Gene X using shRNA expressed from a plasmid
Figure 1: Expression Level of Gene X
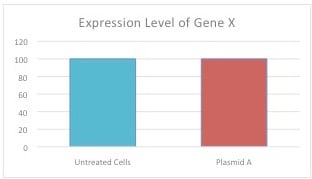
The results shown above are from a single experiment wherein Plasmid A (encoding shRNA against human Gene X in backbone Y) was transfected into human cells.
From this result one could simply conclude that the shRNA didn’t work, as the expression level of Gene X in cells treated with Plasmid A is similar to the expression level of Gene X in the untreated cells—but can we be certain? Do we know whether the plasmid delivered into the cells successfully? Is the shRNA target correct? Is the purified plasmid DNA viable? Or cytotoxic? How and where did things go wrong? The use of proper control plasmids would address and answer these questions.
Types of control plasmids
Part of planning your experiment includes determining what factors need to be controlled for in order to eliminate any alternative interpretation of the results. Typically, plasmid-based experiments employ transfection, negative, positive, and replicate controls.
Transfection controls: empty vector and internal control
A transfection control measures transfection efficiency and enables observation of any effects of the transfection itself (i.e., the vector used in transfection, transfection reagent, or transfection process) may have on the target cells. One transfection control is an empty vector control; specifically, the plasmid without the independent variable. Referring back to the experiment associated with Figure 1, the independent variable is the shRNA. Therefore the empty control vector would be Plasmid A sans shRNA, or backbone Y alone. The empty vector control allows you to examine if the transfection reagents or the transfection process itself has any cytotoxic effects on the target cells.
Another type of transfection control is an internal control vector, which measures transfection efficiency. An internal control may be a plasmid that constitutively expresses a reporter protein (e.g., GFP or luciferase) that is either co-transfected with the test plasmid or transfected into a separate well of your cells. Regardless, the amount of reporter protein activity correlates to both the amount of DNA transfected into the cells and the ability of the cells to express the protein.
In analysis of the result in Figure 1, an internal control, such as the GFP-expressing Plasmid B, could demonstrate whether the cells were transfected successfully and expressing the protein. For example, fluorescence microscopy images resulting from our experiment that includes the aforementioned internal control and is consistent with the result in Figure 1 could look like this:
Figure 2: Expression of Plasmid B (as internal control)
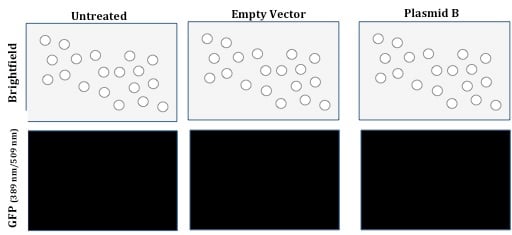
This result indicates that the transfection was not successful due to the absence of GFP fluorescence from the living, viable cells. This result presents an opportunity for troubleshooting and optimization of the transfection reagents and process. It is important to note that optimal experimental conditions, including how much plasmid DNA to use for any individual or co- transfection, should be determined empirically.
Negative controls: untreated cells, empty vector control, and non-targeting control
Negative control conditions and plasmids should produce a null effect (i.e., no phenomenon is observed). In any plasmid-based experiment, untreated cells should be included as these provide the baseline/standard against which other samples can be compared.
The Empty Vector Control (mentioned above) could also serve as an important negative control. In this context, the empty vector control shows any effect of the vector/backbone itself on gene expression in your target cells.
In experiments employing gene targeting or genome editing technologies, such as RNAi or CRISPR, non-targeting controls may be appropriate as they allow you to assess the specificity of your observed result. Non-targeting controls are negative controls that produce a similar product, but do not target an endogenous gene in your experimental cells. For example, in the experiment above, Plasmid A contains an shRNA that targets human Gene X and human cells are being transfected. A proper non-targeting control in this experiment could be an shRNA—in the same backbone as Plasmid A—that does not target any mammalian gene. This non-targeting control is critical to the correct interpretation of the results because it provides an important reference point when analyzing the specificity of the shRNA targeting human Gene X. The non-targeting control also assesses any effects of the general introduction of shRNA into your target cells.
Positive controls
Positive control plasmids should produce the expected phenomenon. One example of a positive control, the internal control vector, was described earlier. Once you are sure your conditions are conducive to the successful delivery of plasmid DNA into the cells, the internal control vector then serves as a positive control for transfection because it produces the expected effect, which is green fluorescent cells (Figure 3).
Figure 3: Expression of Plasmid B (as positive control)
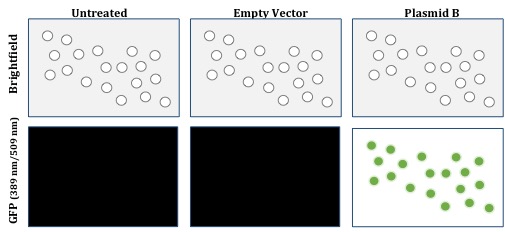
Other positive controls are specific to the experiment and should be designed accordingly. If you are trying activate a gene, you should design a control that shows maximal activation. Likewise if you are trying to repress a gene, your control might be a system where expression of that gene is knocked out completely. These controls may or may not be plasmid-based depending on the experimental needs. Using our experiment as an example, the expected result of the shRNA targeting human Gene X in Plasmid A is the decreased expression of Gene X. Ergo the positive control(s) should decrease the expression of Gene X.
Since the key feature of a control plasmid is to minimize the effects of the non-independent variables within an experiment, both the design and selection of the positive control plasmids should be highly specific to the experiment and the interrogation of the independent variable.
Replicate controls: technical and biological
Reliable results are reproducible from properly controlled experiments. The observed phenotype associated with the independent variable should be consistent over time and from different preparations of the purified test plasmid expressing the independent variable. There are two types of replicate controls: technical and biological.
In general, technical replicates can be thought of as "plate controls". They are NOT independent and are typically derived from one source. Conversely, biological replicates ARE independent and can be thought of as "reproducibility controls". Biological replicates are what makes up your sample size (aka your n value)" and should come from multiple, independent sources. We'll describe these a bit more below, but the papers cited in the reference section provide more in depth information.
Technical replicates
An example of a technical control is the transfection of multiple separate wells (within the same plate) with purified plasmid from the same aliquot/preparation. The replicate control isn’t measuring or assessing reproducibility of the effect of the independent variable because the purified plasmid, cells, and media used in each of the wells are not independent: they were derived from the same source and were incubated on the same plate at the same time. Although the replicate control is important, it speaks to the consistency of the reagents and hands performing the experiment, not the reproducibility or consistency of the observed phenotype associated with the independent variable.
Biological replicates
In any given set of experiments, the biological controls are more time consuming and ultimately more important. Ideally, each biological replicate should use fresh media, test independent aliquots of cells and plasmids, be performed on different days, etc, but outside constraints may impose some limitations and you should be cognizant of these when interpreting your results. The key to biological controls is independence: at the very least you should repeat your whole experiment from start to finish multiple times and on different days. Using our example above, we would test different preparations of Plasmid A in different aliquots of cells on several different days. Biological replicates control for the reproducibility and consistency of the observed phenotype associated with the independent variable and helps ensure the phenotype isn’t singular or associated with only one aliquot/preparation of the test plasmid.
Let us now revisit our experiment. In Figure 1, it appeared as though the shRNA did not knock down expression of Gene X but, as shown in Figure 2, this was likely due to the original transfection conditions. Now that we have successfully transfected our cells (Figure 3), we can continue with our experiment, incorporating additional positive and negative controls, performing multiple replicates, and ultimately getting interpretable results:
Figure 4: Expression Level of Gene X
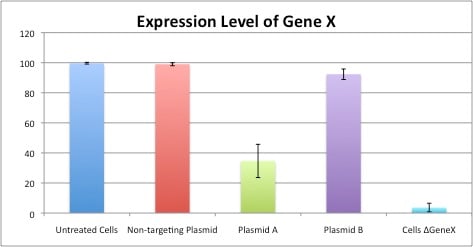
The design and selection of proper experimental controls is not a trivial endeavor, as biological systems have many variables. However daunting designing and executing these steps may be, proper controls are a basic tenet of responsible scientific inquiry and investigation.
References:
1. The problem of pseudoreplication in neuroscientific studies: is it affecting your analysis? Lazic SE. BMC Neurosci. Jan 14;11:5. (2010) PubMed PMID: 20074371. PubMed Central PMCID: PMC2817684.
2. Replicates and repeats—what is the difference and is it significant? Vaux DL, et al. EMBO Rep. Apr; 13(4): 291–296. (2012). PubMed Central PMCID: PMC3321166.
Resources on the Addgene Blog
- Origin of Replication
- The Promoter Region – Let's Go!
- Browse All of Our Plasmids 101 Posts
Topics: Plasmids 101, Plasmids






Leave a Comment