If you’re been following Addgene on #souptwitter or if you just enjoy a warm meal on a winter day, you should be happy to learn that many DNA-based reagents are named for soup ingredients! From pSOUP to mCherry, satisfying ingredients reagents permeate the molecular biologist’s kitchen lab - and scientists using aptamers might know this best, as most fluorescent aptamers are named for a fruit or vegetable!
What is an RNA aptamer?
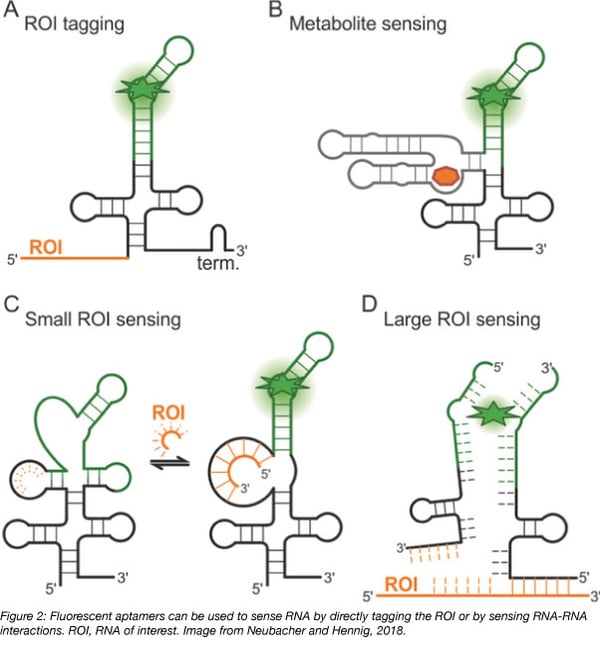
RNA is not inherently fluorescent and can be challenging to track intracellularly, but aptamers permit the study of intracellular RNA dynamics. Aptamers are 10-100 base nucleic acid oligos that bind with high affinity to small molecules to induce their fluorescence. Until a binding event occurs, neither the aptamer nor the small molecule are strongly fluorescent but the binding of an aptamer to its target small molecule activates the fluorescence of the small molecule. Fluorescent RNA aptamers are used to track cellular processes by transforming cells with plasmids encoding aptamer-tagged RNA and later introducing the small molecule into the cell. The fluorescence that is generated allows for qualitative and quantitative measurement of processes including RNA movement by tagging RNA sequences with an aptamer and RNA transcription by placing the aptamer downstream of a known promoter.
Addgene’s Plasmids 101: Aptamer Fluorophores describes the enrichment process used to evaluate oligonucleotides for binding affinity to a specific target, and the molecular processes that make aptamer biology a powerful tool. In this post, we’ll take a deeper dive into the content of Addgene’s aptamer soup by exploring the development and applications of fluorescent RNA aptamers in Addgene’s collection.
 Green fluorescent aptamers
Green fluorescent aptamers
Spinach and Broccoli and their derivatives are aptamers able to bind to a small molecule derivative of GFP called 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI). DFHBI is membrane permeable and non toxic to cells. Because it rapidly photobleaches, DFHBI can only be used for qualitative, but not quantitative, studies. Additionally, Spinach, the first aptamer identified to bind DFHBI, is highly unstable in vivo due to poor folding and thermal instability at 37°C (Ouellet et al., 2016) and requires a tRNA scaffold to improve its stability in vivo.
However, Spinach has proven useful in the engineering of riboswitches. Riboswitches are naturally occurring regions in the 5’ UTR of mRNA that bind metabolites, leading to conformational changes that control the expression of coding RNA. Spinach-based riboswitches can be used as stable metabolite sensors for imaging in live cells. By placing Spinach downstream of a riboswitch, metabolite binding to the riboswitch instead activates Spinach fluorescence.
In the search for something more stable for quantitative imaging, Ming Hammond’s group created Spinach2 using site-directed mutagenesis of Spinach. In doing so, they identified the nucleotides that contribute to the instability of Spinach, and were able to develop an aptamer that was more thermally stable, exhibited less misfolding, and was consequently brighter than Spinach.
The initial studies using Spinach2 showed its ability to track RNA dynamics in a model of Fragile-X tremor/ataxia syndrome (Strack et al., 2013). Since then, Spinach2 has been used to investigate intracellular viral DNA dynamics in neuronal cells (Nilaratanakul et al., 2017) and levels of the second messenger cyclic di-AMP in Gram-positive bacteria (Kellenberger et al., 2013).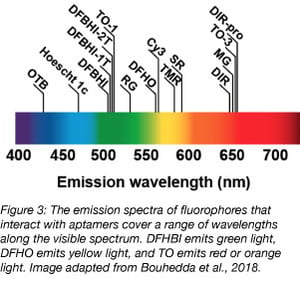
Other variants of Spinach include Baby Spinach, a miniaturized version of Spinach that retains similar levels of fluorescence when bound to DFHBI despite being approximately half the length (Warner et al., 2014); its length reduces artefacts during live cell imaging. Bunch of baby spinach, or BoBS, contains tandem repeats of Baby Spinach. It was developed in a study of artificial guide RNAs that can be used to simultaneously target cellular RNA to specific intracellular locations using Cas9 and track the localization of the complex.
Broccoli, the other green fluorescent aptamer, was identified through directed evolution. First, a pool of RNAs that activate the fluorescence of DFHBI were identified using systematic evolution of ligands by exponential enrichment (SELEX). After 4-6 rounds of SELEX, the RNA pool was cloned into bacterial expression vectors to create a library. Bacteria were transformed, and then sorted by fluorescence-activated cell sorting (FACS) in the presence of DFHBI to identify aptamers capable of inducing the brightest fluorescence (Filonov et al., 2014).
Broccoli is twice as bright as Spinach2 when bound to DFHBI. It’s also smaller than any form of Spinach - only 49 nucleotides in length - and is better able to maintain its fluorescence intracellularly due to a lower dependency on magnesium and higher thermal stability, making it a potentially more useful tool for in vivo studies. Dimerization of Broccoli increases its fluorescence by about 1.8-fold, and may be a better choice for imaging than the monomer. Broccoli has been developed into a split aptamer that can be used to investigate RNA assembly. The split aptamer is composed of a “Top” and “Bottom” portion, each fused to a separate piece of RNA. Fluorescence is observed when these RNAs complex and the entire aptamer can hybridize and fold. (Alam et al., 2017).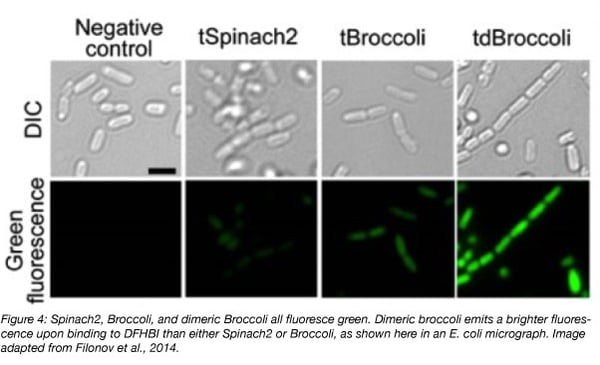
Yellow fluorescent aptamers
Just as vegetables come in a variety of colors, so do aptamers! Corn was recently described in 2017 (Song et al., 2017). Corn naturally dimerizes, and this dimer activates the fluorescence of 3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime (DFHO), a fluorophore modified from DsRed. DFHO shares many structural properties with DFHBI, but its structure allows for greater contact with its RNA aptamer. This increased interaction between Corn and its small molecule makes Corn more photostable than Spinach and Broccoli, and useful for quantitative studies. Specifically, Corn was developed to quantify RNA transcription by RNA Polymerase III. By expressing Corn under promoters for transcripts uniquely transcribed by RNA Polymerase III (5S, tRNA and U6) and measuring Corn fluorescence by flow cytometry, the Jaffrey group was able to quantify the effect of mTOR inhibition on the activity of RNA Polymerase III.
Red and orange fluorescent aptamers
Corn was described in the same study as Red Broccoli and Orange Broccoli, two more aptamers that bind DFHO and fluoresce in the red and orange channels, respectively. Both Red Broccoli and Orange Broccoli are derived from Broccoli, and share similar properties to Broccoli despite binding to a different small molecule. Notably, like Broccoli, Red Broccoli and Orange Broccoli are not photostable, and were presented in the study to support the uniqueness of the photostability of the Corn-DFHO complex.
For dessert (last but not least), the only fruit-named aptamer, Mango, binds to multiple derivatives of thiazole orange (TO) and fluoresces in the orange or red channel, depending on the structure of the TO derivative (Dolgosheina et al., 2014). While 10 derivatives of TO exist, Mango is most frequently used with TO1-biotin or TO3-biotin (the addition of biotin allows for the purification of specific RNAs from cells using this tag, and both TO1-biotin and TO3-biotin contain an acetate modification that reduces nonspecific binding with RNA by about 7x). Mango fluoresces nearly as brightly as Spinach, but binds with much higher affinity to its cognate small molecule. Mango was initially shown to be effective for imaging single molecules and for live-cell imaging in C. elegans. Other groups have used Mango to study RNA-protein complexes (Panchapakesan et al., 2017) and short non-coding RNAs (Autour et al., 2018). Three variants of Mango, coined Mango II, III, and IV, were then identified through microfluidic-assisted in vitro compartmentalisation for their high binding affinity to TO1-biotin. Two of these variants exhibit increased brightness compared to Mango, and all are photostable and useful for both live and fixed cell imaging.
New ingredients in the aptamer soup
In addition to these established RNA aptamer fluorophores, more are in development, including Carrot and Radish from the lab that developed Corn. Aptamers using unnatural nucleotides are also being developed which could help aptamers better bind their target molecules. With more tools comes more possibility for scientific advancement, and in a field with as much potential for growth as RNA biology, we are looking forward to reports on the next additions to aptamer soup!

References
Alam, Khalid K., et al. "A fluorescent split aptamer for visualizing RNA–RNA assembly in vivo." ACS synthetic biology 6.9 (2017): 1710-1721. PubMed PMID: 28548488. PubMed Central PMCID: PMC5603824.
Autour, Alexis, et al. "Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells." Nature communications 9.1 (2018): 656. PubMed PMID: 29440634. PubMed Central PMCID: PMC5811451.
Dolgosheina, Elena V., et al. "RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking." ACS chemical biology 9.10 (2014): 2412-2420. PubMed PMID: 25101481.
Filonov, Grigory S., et al. "Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution." Journal of the American Chemical Society 136.46 (2014): 16299-16308. PubMed PMID: 25337688. PubMed Central PMCID: PMC4244833.
Kellenberger, Colleen A., et al. "RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP." Journal of the American Chemical Society 135.13 (2013): 4906-4909. PubMed PMID: 23488798. PubMed Central PMCID: PMC3775879.
Nilaratanakul, Voraphoj, Debra A. Hauer, and Diane E. Griffin. "Development and characterization of sindbis virus with encoded fluorescent rna aptamer spinach2 for imaging of replication and immune-mediated changes in intracellular viral rna." Journal of General Virology 98.5 (2017): 992-1003. PubMed PMID: 28555544.
Ouellet, Jonathan. "RNA fluorescence with light-up aptamers." Frontiers in chemistry 4 (2016): 29. PubMed PMID: 27446908. PubMed Central PMCID: PMC4923196.
Panchapakesan, Shanker Shyam S., et al. "Ribonucleoprotein purification and characterization using RNA Mango." RNA23.10 (2017): 1592-1599. PubMed PMID: 28747322. PubMed Central PMCID: PMC5602116.
Song, Wenjiao, et al. "Imaging RNA polymerase III transcription using a photostable RNA–fluorophore complex." Nature chemical biology 13.11 (2017): 1187. PubMed PMID: 28945233. PubMed Central PMCID: PMC5679246.
Strack, Rita L., Matthew D. Disney, and Samie R. Jaffrey. "A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat–containing RNA." Nature methods 10.12 (2013): 1219. PubMed PMID: 24162923. PubMed Central PMCID: PMC3852148.
Warner, Katherine Deigan, et al. "Structural basis for activity of highly efficient RNA mimics of green fluorescent protein." Nature structural & molecular biology 21.8 (2014): 658. PubMed PMID: 25026079. PubMed Central PMCID: PMC4143336.
Additional resources on the Addgene blog
- Learn more about aptamer fluorophores
- Investigate other fluorescent biosensors
- Find other uses for RNA aptamers
Resources at Addgene.org
- Browse fluorescent protein plasmids and resources
- See all Jaffrey lab plasmids
Topics: Fluorescent Proteins, Other Fluorescent Protein Tools





Leave a Comment