Addgenie Mary Gearing contributed to the content of this article.
Biosensors (‘biological sensors’) are biological tools that monitor a process or detect a given molecule. The sensor component is usually a protein that undergoes a conformational change in response to the molecule it detects. This change then generates a reporter signal. Reporter signals may be electrochemical or light-based, with luminescent and fluorescent reporters being especially popular. We’ll give you an introduction to fluorescent biosensors, but keep in mind that there is a lot of variety in how biosensors work, and you should always check the associated publication for the specifics of your chosen plasmid.

Types of biosensors
When choosing a biosensor, you first need to decide the best way to track or measure the process you’re interested in. Depending on the process you would like to monitor, there may be multiple steps you could examine, i.e. for an enzymatic reaction, you could measure consumption of a substrate OR production of a product OR enzyme activity.
Small molecule biosensors
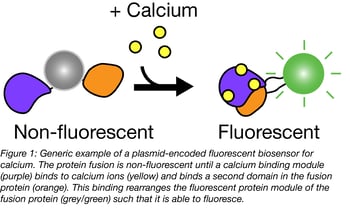 Addgene has many sensors that measure the concentration of small molecules like arabinose or hydrogen peroxide or ions like calcium and zinc. Some biosensors contain a single fluorescent protein fused to a domain that senses the concentration of a given ion - we’ll use calcium biosensors as an example. Many calcium biosensors include GFP fused to calmodulin (CaM) and the chicken myosin kinase calmodulin binding region RS20. When calcium is absent, the construct does not fluoresce; when present, calcium induces CaM/RS20 interaction, thereby changing the FP’s structural organization and activating the fluorescent signal. Blue flame GECO biosensors from Robert Campbell’s lab display calcium-dependent increases in fluorescence of up to 11,000-fold.
Addgene has many sensors that measure the concentration of small molecules like arabinose or hydrogen peroxide or ions like calcium and zinc. Some biosensors contain a single fluorescent protein fused to a domain that senses the concentration of a given ion - we’ll use calcium biosensors as an example. Many calcium biosensors include GFP fused to calmodulin (CaM) and the chicken myosin kinase calmodulin binding region RS20. When calcium is absent, the construct does not fluoresce; when present, calcium induces CaM/RS20 interaction, thereby changing the FP’s structural organization and activating the fluorescent signal. Blue flame GECO biosensors from Robert Campbell’s lab display calcium-dependent increases in fluorescence of up to 11,000-fold.
FRET biosensors are another common class of biosensors. Blue flame “cameleon” calcium sensors created by Roger Tsien’s lab contain two fluorescent proteins (a FRET pair) separated by CaM and M13. When calcium binds, it triggers CaM/M13 interaction that increases FRET transfer between the fluorescent proteins. FRET is highly amenable to biosensor creation, and custom FRET biosensors can be created using the cpFRET kit created by Olivier Pertz’s lab.
Other biosensors track cellular characteristics like pH, redox state, and voltage across membranes. These sensors generally follow the same pattern described above - a sensing domain induces changes in a fluorescent signal based on ligand presence or changing cellular conditions.
Other types of biosensors
To focus on a particular gene or protein, check out our gene-specific biosensors. These constructs are often used to monitor enzymatic activity or phosphorylation over time. You can also use biosensors to follow processes such as protein or DNA binding, enzyme or transcriptional activation, conformational changes, translation, and protein translocation.
Targeting your biosensor
Before expressing a biosensor in eukaryotic cells, think about the best place for the biosensor to be expressed. Are you looking at an intracellular or extracellular molecule? By fusing a biosensor to an organelle-specific targeting signal or a transmembrane domain, biosensors can be targeted to a particular cellular compartment such as the nucleus, mitochondria, endoplasmic reticulum (ER), vacuole or cellular membranes. Examples include blue flame mitochondria-targeted calcium sensor CMV-mito-GEM-GECO1 and blue flame extracellular glutamate sensor pCMV(MinDis).iGluSnFR.
Picking your reporter
Don’t forget about the other end of the biosensor: which fluorescent protein is most compatible with your lab and experimental setup? Consider what kind of instrumentation you have available, and the conditions of your experiment. For example, does the fluorescent tag need to be pH stable or withstand a particular solute concentration or temperature? For further advice, see our posts on how to select a fluorescent protein for single and multi-color imaging experiments.
Finally, what kind of system will you use to detect your fluorescent protein? The type of detection method depends on what type of information you are looking for. Below we have summarized some of the common fluorescent detection methods and the types of experiments they are commonly used for.
Table 1. Points to Keep in Mind When Performing Biosensor Experiments
| Experiment | Detection Method | Notes |
| Quantification of a process | Plate-based assays (plate reader) |
Detect the average amount of fluorescence across a population of cells (i.e. in a plate well). Use to track a process (e.g. enzyme activity) over time or under various conditions |
|
Localization within cells |
Fluorescence microscopy |
Visualize components of individual cells |
| Separation/identification of fluorescently-labeled components | Fluorescence-activated cell sorting (for cells); fluorescent HPLC (proteins or other molecules) | Isolate or quantify cells, proteins or other molecules that have been labelled with your fluorescent molecule |
Additional Resources on the Addgene Blog
- Check out the Rosella Autophagy Sensor
- Read about Aptamer Fluorophores
- Use CRISPRainbow for Genome Visualization
Resources on Addgene.org
- Find Biosensor Plasmids
- Find Optogenetics Plasmids
- Visit the Fluorescent Protein Guide Pages
Topics: Fluorescent Proteins, Fluorescent Proteins 101, Fluorescent Biosensors





Leave a Comment