What is an aptamer?
Nearly 30 years ago, two independent groups, led by Jack Szostak and Larry Gold, developed methods for selecting and amplifying RNA sequences that could bind very specifically to target molecules. Using a technique called systematic evolution of ligands by exponential enrichment (SELEX), some 1010 oligonucleotides could be screened for their affinity to a wide range of non-nucleotide targets. These RNA molecules, which could bind their targets with high specificity and affinity, were eventually called aptamers, from the Latin aptus, meaning “to fit”. SELEX could be used to classify DNA aptamers as well, and over the course of the next two decades, these nucleotide-based ligand binders would prove to be highly adaptable tools.
The key to aptamers’ flexibility is their broad range of targets, including proteins, peptides, amino acids, drugs, metal ions, and cells (even pathogenic bacteria). That range is possible because aptamers can either fold to incorporate their target molecule, such as a metal ion, or fold to incorporate themselves into a larger target, such as a protein. Such flexibility makes aptamers ideal candidates for biosensors. Aptamer biosensors are used in a variety of applications, including detection of environmental pollutants, drug discovery, and disease diagnosis.
As biosensors, aptamers hold a few important advantages over their amino acid-based counterparts, antibodies and enzymes, such as:
- Design and selection for targets can be done largely in vitro;
- Easy and reproducible commercial synthesis (those of us who have used antibodies for diagnostics know how frustrating it can be to get a “bad batch”);
- Customizable sensitivity and detection - since they frequently undergo conformational changes after target binding, aptamers are highly customizable in terms of how they actually “report” as biosensors.
Making aptamers glow
The flexibility of aptamers means they can be used with or as fluorophores in several different ways. Researchers have used oligonucleotides in fluorescent visualization techniques for decades, most notably for fluorescent in situ hybridization (FISH) where fluorophore-linked antibodies are used to visualize RNA. Low signal-to-noise ratios with such techniques can be problematic, however. Well designed aptamers can help reduce such background, thereby improving traditional visualization techniques. In addition, their incredible versatility opens aptamers for use in many entirely new applications including aptamer based fluorescent biosensors.
Aptabeacons
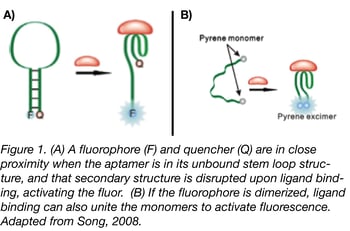 Fluorescence resonance energy transfer (FRET) technology has been studied and used in assays for over half a century; the Addgene blog has an excellent primer and tips from experts on FRET techniques. By combining standard donor/acceptor FRET know-how with aptamers, aptabeacons (or aptatags) were born. Aptabeacons can be created in many different ways, but the general premise is that an aptamer in its unbound state contains a fluorophore and a quencher in close proximity. For instance, the fluorophore and quencher may form the base of the stem in an RNA stem-loop structure (see Fig. 1a). When the aptamer binds its ligand, the stem loop is disrupted, removing the quencher from the proximity of the fluorophore and subsequently leading to a positive signal. The same effect can be achieved for a fluorophore that relies on dimerization for activation, only in this case ligand binding would bring the relevant ends together rather than separate them (Fig. 1b). One can also make “signal-off” biosensors in which the aptamers fluoresce until they bind to the appropriate ligand, but relying on a lack of fluorescence for readout may render this approach less sensitive in most applications.
Fluorescence resonance energy transfer (FRET) technology has been studied and used in assays for over half a century; the Addgene blog has an excellent primer and tips from experts on FRET techniques. By combining standard donor/acceptor FRET know-how with aptamers, aptabeacons (or aptatags) were born. Aptabeacons can be created in many different ways, but the general premise is that an aptamer in its unbound state contains a fluorophore and a quencher in close proximity. For instance, the fluorophore and quencher may form the base of the stem in an RNA stem-loop structure (see Fig. 1a). When the aptamer binds its ligand, the stem loop is disrupted, removing the quencher from the proximity of the fluorophore and subsequently leading to a positive signal. The same effect can be achieved for a fluorophore that relies on dimerization for activation, only in this case ligand binding would bring the relevant ends together rather than separate them (Fig. 1b). One can also make “signal-off” biosensors in which the aptamers fluoresce until they bind to the appropriate ligand, but relying on a lack of fluorescence for readout may render this approach less sensitive in most applications.
Learn About Fluorescent Biosensors
Light-up aptamers (or, fluorescing with veggies)
Though the aptabeacons can be used as powerful biosensors, the bound protein fluorophores and quenchers are still relatively bulky. One of the advantages of oligonucleotides over proteins is their modularity, which is lost as soon as the aptamer is bound to a protein fluor. Transcribing and translating a protein fluorophore also takes time - bypassing translation altogether with an oligonucleotide-only sensor could save precious minutes in a time sensitive detection assay.
In 1999, Grate and Wilson paired laser cleavage and an RNA aptamer that bound the compound malachite green (MG) to create a non-protein based visualization technique. Ultimately, MG proved to be too toxic for many in vivo applications, but this work proved the concept that RNA could be used to induce fluorescence outside the context of a paired fluorophore protein. Years later, Samie Jaffrey’s laboratory took advantage of their deep knowledge of the nature of GFP fluorescence to create an improved fluorescent aptamer system. Knowing that GFP’s structure stabilizes a 4-hydroxybenzylidene imidazolinone (HBI)-like compound allowing it to fluoresce, the Jaffrey lab asked: Could an RNA aptamer do the same thing?
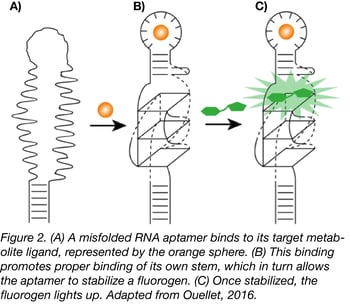 Using the HBI derivative 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) with relatively low fluorescence in vivo, the Jaffrey lab used SELEX to find a 98 nt RNA aptamer that would bind the fluorogen and increase its fluorescence, essentially mimicking GFP. Though the aptamer, called Spinach, required longer exposure time than its protein equivalent and was less stable due its RNA nature, it paved the way to new biosensor possibilities. For instance, further selection of Spinach with a target metabolite creates an aptamer that will only bind to DFHBI in the presence of that metabolite (Fig.2).
Using the HBI derivative 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) with relatively low fluorescence in vivo, the Jaffrey lab used SELEX to find a 98 nt RNA aptamer that would bind the fluorogen and increase its fluorescence, essentially mimicking GFP. Though the aptamer, called Spinach, required longer exposure time than its protein equivalent and was less stable due its RNA nature, it paved the way to new biosensor possibilities. For instance, further selection of Spinach with a target metabolite creates an aptamer that will only bind to DFHBI in the presence of that metabolite (Fig.2).
Spinach eventually evolved into Spinach2, a more stable and brighter version of the aptamer, but it still had limitations in vivo. Using a combination of SELEX and fluorescence-activated cell sorting (FACS) of E. coli transformed with aptamer libraries, the Jaffrey group selected for a 49-nucleotide aptamer they called Broccoli. As a product of SELEX and FACS, Broccoli is inherently more stable in cells. The new aptamer also has a higher melting temperature than its Spinach ancestors, and has a lower magnesium dependence. Though initially still unstable in eukaryotes, the Jaffrey lab has since developed new RNA-based scaffolds to contain and stabilize Broccoli cassettes, thereby extending their use into mammalian cells. They’ve also dimerized Broccoli to further enhance its biosensor capabilities (see Fig. 1b).
The future of aptamer fluorophores
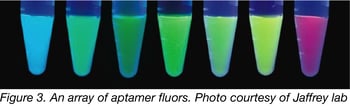 The Jaffrey lab and others continue to hone their selection techniques to come up with new colors (Fig. 3), such as Radish and Carrot, and further means to increase aptamer stability and decrease background in vivo. The nature of aptamers makes them excellent candidates for developing assays for otherwise difficult-to-study RNA-modifying enzymes, such as the RNA demethylases FTO and ALKBH5. Their ease of production have also made aptamers an important component of high throughput assays with microarrays and biochips. Ultimately, there are just some applications that lend themselves to aptamer-based instead of protein-based fluorophores, including:
The Jaffrey lab and others continue to hone their selection techniques to come up with new colors (Fig. 3), such as Radish and Carrot, and further means to increase aptamer stability and decrease background in vivo. The nature of aptamers makes them excellent candidates for developing assays for otherwise difficult-to-study RNA-modifying enzymes, such as the RNA demethylases FTO and ALKBH5. Their ease of production have also made aptamers an important component of high throughput assays with microarrays and biochips. Ultimately, there are just some applications that lend themselves to aptamer-based instead of protein-based fluorophores, including:
- Environments with higher temperatures, since proteins can denature while aptamers are more stable and can go through cycles of denaturation/renaturation.
- In vivo assays in which protein fluors are likely to trigger immunoresponses. Nucleotides, in general, are far less immunogenic than proteins.
- Assays that require recognition of ions or particularly small molecules that do not generate an immune response. Aptamers can be selected for these small ligands.
References
1. Ma, Hanhui, et al. “Multicolor CRISPR labeling of chromosomal loci in human cells.” Proc. Natl. Acad. Sci. USA 112(10) (2015): 3002-7. PubMed PMID: 25713381. PubMed Central PMCID: PMC4364232.
2. Baker, Monya. “RNA imaging in situ”. Nature Methods 9, 787–790 (2012).
3. Filonov, Gregory S., et al. “In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies.” Chem Biol. 2015 May 21;22(5):649-60. PubMed PMID: 26000751. PubMed Central PMCID: PMC4441765.
4. Guo, Peixuan AND Haque, Farzin. RNA Nanotechnology and Therapeutics. Boca Raton: CRC Press, 2014.
5. Hayat, Akhtar and Marty, Jean L. “Aptamer based electrochemical sensors for emerging environmental pollutants.” Front Chem. 2014; 2: 41. PubMed PMID: 25019067. PubMed Central PMCID: PMC4071757.
6. Hong, He, et al. “Molecular Imaging with Nucleic Acid Aptamers.” Curr Med Chem. 2011;18(27):4195-205.PubMed PMID: 21838686. PubMed Central PMCID: PMC3205285.
7. Ouellet, Jonathan. “RNA Fluorescence with Light-Up Aptamers.” Front Chem. 2016 Jun 28;4:29. PubMed PMID: 27446908. PubMed Central PMCID: PMC4923196.
8. Sassolas, Audrey, et al. “Optical detection systems using immobilized aptamers.” Biosens Bioelectron. 2011 May 15;26(9):3725-36. PubMed PMID: 21419619.
9. Song, Kyung-Mi, et al. “Aptamers and Their Biological Applications.” Sensors (Basel). 2012; 12(1): 612–631. PubMed PMID: 22368488. PubMed Central PMCID: PMC3279232.
10. Song, Shiping, et al. “Aptamer-based biosensors.” TrAC Trends in Analytical Chemistry 27(2):108-117 · February 2008.
11. Svensen, Nina and Jaffrey, Samie R. “Fluorescent RNA Aptamers as a Tool to Study RNA-Modifying Enzymes.” Cell Chem Biol. 2016 Mar 17;23(3):415-25. PubMed PMID: 26877022. PubMed Central PMCID: PMC4800036.
12. Wang, Rongsheng E., et al. “Aptamer-Based Fluorescent Biosensors.” Curr Med Chem. 2011;18(27):4175-84. PubMed PMID: 21838688. PubMed Central PMCID: PMC3205236.
13. Xiao, Saijin, et al. “Aptamer-based assay for prion diseases diagnostic”. Procedia Environmental Sciences. Volume 12, Part B, 2012, Pages 1348-1353.
Additional Resources on the Addgene Blog
Resources at Addgene.org
- Find the Spinach Plasmids at Addgene
- Find Broccoli Plasmids at Addgene
- View All Fluorescent Protein Resources






Leave a Comment