At Addgene, we love GFP, and we’re always excited when depositors find new ways to make this workhorse protein even more useful! From FPs optimized for oxidizing environments to photoconvertible variants, it seems like GFP is always learning new things. Now, work from Connie Cepko’s lab allow researchers to activate transcription or Cre recombinase activity only in the presence of GFP. These systems, known as T-DDOG and Cre-DOG, respectively, repurpose popular GFP reporter lines for more sophisticated experimental manipulations, saving the time and money needed to develop new lines.
Using nanobodies to create GFP scaffolds for transcriptional activation
The project began with a frustration many researchers have experienced: wanting to ask and answer experimental questions, but not having the tools needed to do so. Cepko lab researcher Jonathan Tang wanted to express a variety of proteins in single cell types in the mouse, but knew it would take years to develop the mouse lines required for this purpose. Tang wondered if he could use previously established mouse lines expressing GFP in specific cell types to do more than just label cells. If GFP could be co-opted to control gene expression, he could then selectively manipulate only GFP-labeled cells.
Once Tang and Cepko found a description of GFP-binding nanobodies, the project really took off. Unlike most antibodies, nanobodies, which occur naturally in camels, consist of a single heavy chain, and they are small and stable inside cells. Nanobodies binding GFP were designed in 2009. Subsequent work showed that nanobodies could be fused to other proteins, and these fusions retained the ability to bind GFP.
The idea of using GFP as a scaffold suddenly seemed very realistic. Envisioning GFP as a substrate promoting dimerization, Tang et al. tested pairs of their GFP-binding proteins (GBPs) to find those that could co-occupy GFP. Once they’d found suitable pairs, they constructed three components: GBPa-VP16 (activation domain); GBPb-GAL4 (DNA-binding domain); and a UAS-driven luciferase reporter construct. In the absence of GFP, no reporter output was observed in their 293 cell culture system. In cells cotransfected with GFP and the GBPs, GFP linked the two GBPs together to create a complete transcription factor, activating luciferase transcription. Tang et al. coined this system T-DDOG (Transcriptional Devices Dependent On GFP).
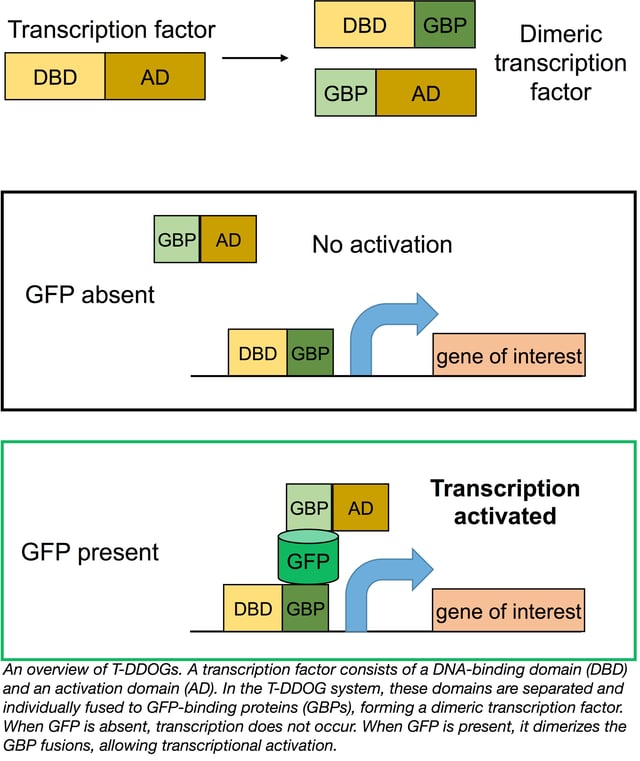
In testing the specificity of their system, Tang et al. found that YFP and CFP can activate certain T-DDOG constructs similarly to GFP, but commonly used red fluorescent proteins dsRed, mCherry, and TdTomato do not induce transcription. Thus, T-DDOGs can be used in combination with green-red Cre-lox systems. T-DDOGs can also be designed with other DNA-binding domains, including the commonly used LexA and rTetR systems such that they activate gene expression from different promoters.
Moving to an in vivo system, Tang et al. electroporated T-DDOGs, GFP, and a reporter construct into the mouse retina. GFP expression robustly activated the reporter gene TdTomato, whose expression was absent without electroporated GFP. They then tested T-DDOGs with two mouse GFP reporter lines; again, TdTomato was seen only in cells with GFP, at a high activation frequency of 56-93%. In the converse test, 98% of TdTomato expressing cells were GFP+, indicating a robust, but specific system. T-DDOGs also successfully regulated expression of channelrhodopsin-2, commonly used in optogenetics, opening up the possibility of adapting GFP lines for optogenetics experiments in specific cell populations.
Listen to Our Podcast Interview with Connie Cepko
Combining Cre and GFP with Cre-DOGs
The T-DDOG results showed that GFP could regulate transcription in a cell-type specific manner, and Tang and Cepko were eager to see if they could apply GFP regulation to other types of proteins. They started with another biological workhorse, Cre recombinase, which induces recombination at LoxP sites. Placing a lox-STOP-lox cassette upstream of a gene of interest blocks transcription when Cre is absent. When Cre is present, it removes the STOP cassette, activating gene expression.
Although Cre isn’t a modular protein, Tang et. al were able to create a split version of Cre that is only active when dimerized by the GBPs/GFP. The new system, CRE-DOG (Cre Dependent On GFP), is activated by GFP and derivatives GFP and YFP, but not red fluorescent proteins, as seen with T-DDOGs.
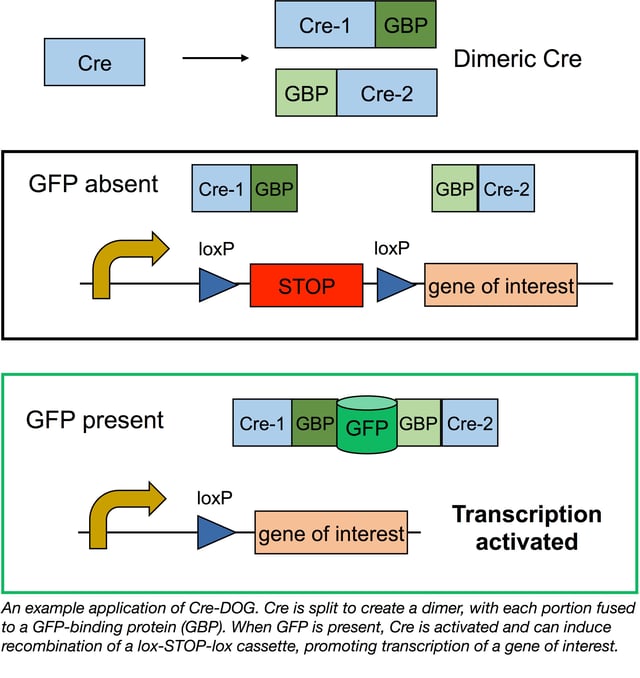
Like T-DDOGs, Cre-DOG is both robust and specific. When tested in retinal electroporation studies, DsRed reporter expression was induced in ~76% of GFP+ cells, and 100% of DsRed+ cells also expressed GFP. Using AAV constructs, Tang et al. found that Cre-DOG could be delivered throughout the nervous system, including the motor cortex, cerebellum, and spinal cord. Cre-DOG is also suitable for optogenetic studies, and since cells infected with AAV-Cre-DOG retain normal neuronal function, this system may make optogenetics in specific cell populations even easier.
Advantages and other possibilities
It’s clear that Cre-DOG and T-DOGGs open up many new possibilities for GFP-labeled lines, and they each have their own advantages. T-DOGGs can be made drug-inducible using the rTetR system, but Cre-DOG lacks the toxicity that can be seen with high levels of transcription activation domains. Both systems are easily adaptable to neuroscience applications, including optogenetics, and they should make it much easier to conduct functional studies in the nervous system.
More broadly, Cepko and Tang note that their scaffold system is likely applicable to many types of proteins. For proteins with a modular structure, constructing split variants should be relatively straightforward, but it is also possible to create split versions of non-modular proteins like Cre. Could Cas9-DOG be developed soon to allow genome modification only in a small subset of cells? Other proteins may also be utliized as scaffolds, including the set of red fluorescent proteins.
The T-DOGGs and Cre-DOG are available from Addgene, and we’re eager to see how you use them in your research!
References
1.Tang, Jonathan C.Y., et al. “A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation.” Cell 154(4) (2013): 928-939. PubMed PMID: 23953120. PubMed Central PMCID: PMC4096992.
- Find plasmids from this publication at Addgene.
2. Tang, Jonathan C.Y., et al. “Cell type-specific manipulation with GFP-dependent Cre recombinase.” Nat. Neurosci. 18(9) (2015): 1334-1341. PubMed PMID: 26258682.
- Find plasmids from this publication at Addgene.
3. Kirchhofer, A., et al. “Modulation of protein properties in living cells using nanobodies.” Nat. Struct. Mol. Biol. 17 (2010): 133–138. PubMed PMID: 20010839.
4. Caussinus, E., Kanca, O., and Affolter, M. “Fluorescent fusion protein knockout mediated by anti-GFP nanobody.” Nat. Struct. Mol. Biol. 19 (2012), 117–121. PubMed PMID: 22157958.
Addgene Resources
Topics: Fluorescent Proteins, Cre-lox, Other Fluorescent Protein Tools






Leave a Comment