This post was originally written by Joel McDade and significantly updated in 2022 by Susanna Stroik.
The advent of CRISPR/Cas9 has made it easier than ever to make precise, targeted genome modifications. Cas9 has been modified to enable researchers to knock out, knock in, base edit, activate, repress, and even image your favorite gene. With so many Cas9 reagents available, it can be difficult to decipher which one is right for your unique experiment. Here, we will introduce you to the wide array of Cas9s (many available through Addgene’s repository) and make it easy to find your perfect match.
Choosing a Cas9 for your knockout experiment
Generating a no-fuss genetic knockout
Knock-out cells or animals are engineered by introducing Cas9-mediated frameshift mutations in the gene of interest. The genomic target for Cas9 and the introduction of this mutation can be any site early in the coding region of the gene of interest. This gives you loads of sequences to select from - in many cases several exons’ worth of DNA. Any ∼20 nucleotides of DNA can serve as the target for Cas9’s single guide RNA (sgRNA), provided it meets two conditions:
1) The sequence is unique compared to the rest of the genome.
2) The target is present immediately upstream of a Protospacer Adjacent Motif (PAM), the sequence NGG.
If these conditions apply to your experimental target, SpCas9 and its enhanced variants, eSpCas9 and HypaCas9, are great choices. Check out our SpCas9 containing plasmids, optimized for a variety of species.
Specificity concerns?
If off-target effects are a worry, consider a dual-sgRNA system. This system uses a “nicking” Cas9, in which one of Cas9’s two critical enzymatic residues has been converted to an alanine (D10A or H840A), changing its double-strand cut to a single-stranded nick. In order to produce a double stranded break, two sgRNAs must be used to target two Cas9-nickases (Cas9n) to the target locus, enhancing specificity over a single SpCas9. The drawback of this approach is that two sgRNAs targeting opposite strands must be available in moderate proximity to each other.
If having two sgRNAs isn’t feasible for your experiment, ensure your sgRNA sequence has little homology to other regions of the genome and consider using a Cas9 variant with higher specificity (eSpCas9 and HypaCas9) instead.
Choosing a Cas9 for site-specific modifications
Site-specific deletions and knockouts
Some experiments may require the introduction of a mutation at a specific site e.g., deleting the catalytic residue of a protein or knocking out a very small peptide. In these cases, you have little flexibility as to where your sgRNA needs to be located and there may not be a NGG PAM site anywhere to be found. If you’re lucky enough to find a SpCas9 compatible PAM at your site of interest, that is the most efficient option. If there is no NGG in your genomic region, then read on for your best options.
There are an expanding number of Cas9s with looser PAM requirements compared to SpCas9. xCas9 has a PAM requirement of NG, GAA, or GAT, making it a more flexible cutter with the added bonus of low off-target effects. The SpG variant, discovered by the Kleinstiver group has a PAM of NGN (Walton et al., 2020). Further, this group also characterized a second Cas9 variant, SpRY, which cuts at NRN PAMs with high efficiency, as well as NYN, but with a slightly lower efficiency. Information on even more alternative PAM Cas9s can be found in “The PAM Requirement and Expanding CRISPR Beyond SpCas9.”
Single base pair edits
Is the location and type of edit you need to introduce highly specific? Perhaps you need to introduce a single nucleotide modification at a defined amino acid to mimic a patient mutation or maybe to disrupt a catalytic residue. Good news! You can achieve your precise edit without even making a break.
Base editors are dead Cas9 (dCas9) or nCas9 enzymes fused to either cytosine or adenosine deaminases to facilitate a single base pair edit. (Cytosine deaminases edit C-to-T and adenosine deaminases edit A-to-G.) In short, the impaired Cas9 localizes the deaminase to where the edit is desired, and it performs the programmed edit within the editing window (bases 4-8 of the protospacer). While this is a small window, this system can be used with a variety of impaired Cas9 proteins (such as SpRY) with altered PAM requirements, giving it more flexibility. A consideration with this tool is that the desired edit will be performed on all relevant bases within the editing window, for example all C’s will edit to T’s. Thus, it’s important that there are no “bystander bases” if you choose this system.
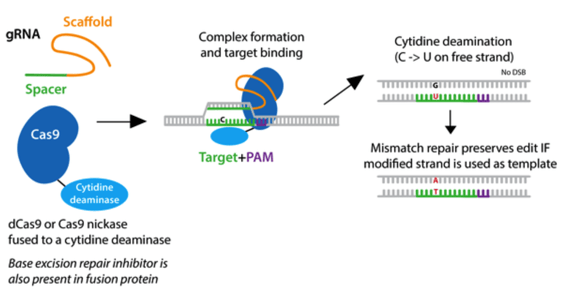 |
| Fig. 1: Cytidine deamination takes place on the free strand of DNA and converts a C to U without producing a double strand break. |
Prime editors are the second generation of base modifiers available. This system utilizes an RNA template containing the desired edit adjoined to the sgRNA, a reverse transcriptase, and a nCas9. While this system has been shown to be less efficient than base editing (Anzalone et al., 2019), it does not require an expertly positioned PAM without undesired bases nearby that may undergo unwanted editing. Just like with base editing, alternative Cas9s can be utilized allowing for PAM flexibility.
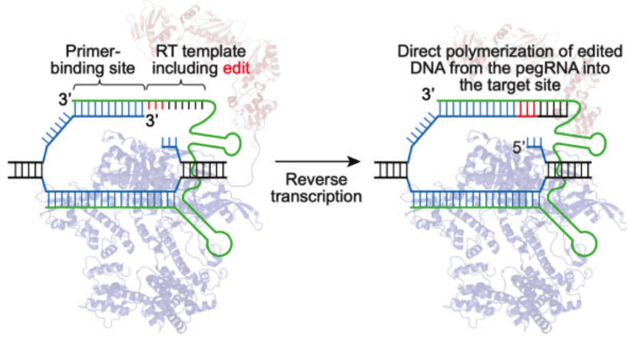 |
| Fig. 2: Prime editor engaging target DNA. The reverse transcriptase polymerizes DNA onto the target strand based on the sequence of the pegRNA. Image from David Liu. |
Activation and repression using Cas9
Don’t want to permanently modify the genome but still want to affect gene expression? dCas9 can be used as a platform to deliver various cargo to a specific DNA locus by directly fusing the cargo to dCas9. This technique has been exploited to localize factors such as transcriptional activators and repressors to target genes.
Repressing target genes using dCas9-based repressors
Early experiments using dCas9 in bacteria demonstrated that targeting dCas9 to transcriptional start sites was sufficient to repress transcription by blocking its initiation. In mammalian cells, robust repression requires targeting dCas9 along with transcriptional repressors to the promoter region of the gene of interest. dCas9-based repressors are useful when knocking out a particular gene isn’t feasible. For example, temporary repression or knockdown allow you to study essential genes whose knockout would cause cell lethality. The Krüppel associated box (KRAB) is a naturally occurring transcriptional repressor domain which is frequently fused to Cas9 for this purpose. dCas9-KRAB plasmids for repressing target genes in a variety of species and cell types can be found on Addgene’s website.
Activating target genes using dCas9-based activators
The simplest of the dCas9 activators consists of dCas9 fused to a single transcriptional activation domain - VP64. Later generations of activators include the SunTag system which facilitates recruitment of multiple activation domains to the same genetic locus via co-expression of an epitope tagged dCas9 and antibody-activator fusion proteins. The Synergistic Activation Mediator complex, generally the strongest activator, consists of a dCas9-VP64 fusion and a modified gRNA that is capable of interacting with an additional RNA-binding transcriptional activator. Plasmids for activating target genes in a variety of species and cell types can be found on Addgene’s website.
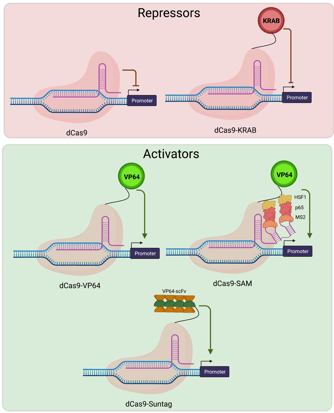 |
| Fig. 3: Cas9 repressors include dCas9 alone and a dCas9-KRAB fusion, both targeted to the promoter region. Cas9 activators include Cas9 fused to VP64, VP64 along with other SAM factors, and the Suntag array of factors, all targeted to the promoter region. |
Ok, I have a Cas9! What do i do next?
Designing or selecting a sgRNA
Addgene may carry a validated sgRNA for your target gene. Validated gRNAs are a great bang for your buck – they have been used successfully in experiments and published in peer reviewed journals. If you need to generate a new sgRNA for your experiment, select from a variety of empty sgRNA vectors and design your sgRNA targeting sequence using one of the many freely available sgRNA design programs.
Find Validated gRNAs for Your Next CRISPR Experiment
Cas9 expression systems
Addgene carries a variety of Cas9 containing plasmids that have been optimized for expression in different species and cell types including (but not limited to) bacteria, yeast, plants, drosophila, worms and mammals (Browse plasmids by model organism, here). More information on the various delivery systems can be found here “CRISPR 101: Mammalian Expression Systems and Delivery Methods”.
Validating your edit
Once you have delivered Cas9 and a sgRNA to your target cells, it is time to confirm that your target sequence has been modified! The exact protocol may vary depending on your specific experiment, but a broad overview of the various ways in which you can verify your genome edit can be found in our blog post entitled “CRISPR 101: Validating Your Genome Edit”.
Is Cas9 not your Cas of choice?
There are plenty of other enzymes to choose from within the Cas family if Cas9 isn’t right for your experiment. Keep an eye out for our upcoming review of these options in our CRISPR 101 series!
References and resources
References
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020 387 38, 824–844 (2020).
Moreno-Mateos, M. A. et al. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat. Commun. 2017 81 8, 1–9 (2017).Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science (80-. ). 368, 290–296 (2020).
More resources on the Addgene blog
- Read all the posts in our CRISPR 101 Series
- Learn about the PAM Requirements of Different Cas9 variants
- Learn about CRISPR Pooled Library Experiments
Additional resources on the Addgene website
- Find CRISPR Plasmids
- Catch up on your CRISPR background with our CRISPR Guide
- Find Software Tools for gRNA Design
Topics: CRISPR, CRISPR 101, Cas Proteins








Leave a Comment