Originally published Nov 12, 2015 and last updated Aug 20, 2020.
Cas9 can be used to modify any desired genomic target provided that (1) the sequence is unique compared to the rest of the genome and (2) the sequence is located just upstream of a Protospacer Adjacent Motif (PAM sequence). The 3-5 nucleotide PAM sequence serves as a binding signal for Cas9 and this sequence is a strict requirement for Cas9-mediated DNA cleavage.
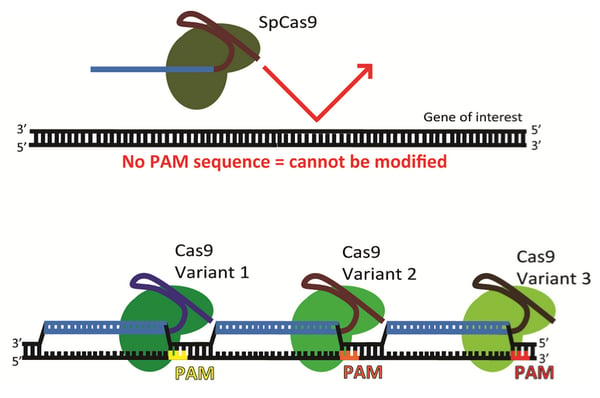
The need for more PAM sequences
While PAM sequences for the commonly used S. pyogenes Cas9 (3'-NGG) are abundant throughout the human genome, they are not always positioned correctly to target a particular gene. Furthermore, a target sequence may have high homology elsewhere in the genome. These off-target sequences may be unintentionally mutated along with the desired target locus.
The PAM sequence is of particular concern when trying to edit a gene using homology directed repair, since HDR-mediated gene editing is most efficient when target sites are located in close proximity to the region to be edited. In this blog post, we will cover three ways to circumvent this limitation: 1) the use of novel S. pyogenes Cas9 variants with varying PAM sequences, 2) the use of Cas9 homologs derived from species other than S. pyogenes, and 3) the use of non-Cas9 enzymes.
(For more details on the PAM site, check out this video from IGI!)
Synthetic S. pyogenes Cas9s with novel PAM recognition
In 2015, Keith Joung’s lab performed a series of positive selection screens in bacteria to identify mutants of S. pyogenes Cas9 that were able to cleave target DNA sequences located upstream of either NGA or NGC PAM sequences (Kleinstiver et al. 2015). From these screens, they identified four novel SpCas9 variants with altered PAM binding specificity:
|
SpCas9 variant |
Mutations (relative to SpCas9) |
PAM sequence |
|
D1135E |
NGG |
|
|
D1135V, R1335Q and T1337R |
NGAN or NGNG |
|
|
D1135E, R1335Q and T1337R |
NGAG |
|
|
D1135V, G1218R, R1335E and T1337R |
NGCG |
The D1135E variant is far more selective for the canonical S. pyogenes PAM sequence NGG compared to wild-type SpCas9, which also displays some cleavage with an NGA PAM. This variant may increase the specificity of genome modifications at DNA targets adjacent to NGG PAM sequences when used in place of wild-type SpCas9. The remaining variants (VQR, EQR, and VRER) recognize novel PAM sequences (shown above). The VQR, EQR, and VRER Cas9 variants are capable of cleaving genomic DNA in mammalian cells and zebrafish embryos, and they can be used to modify genomic loci that cannot be modified using wild-type SpCas9. The number of off-target cleavage events for the VQR and VRER variants is similar to wild-type SpCas9, indicating that the variants are likely just as selective as wild-type SpCas9. These VQR, EQR and VRER SpCas9 variants effectively double the targeting range of CRISPR/Cas9 within the human genome.
Another variant, xCas9 3.7 has 7 mutations found in the REC2, REC3, and PAM interacting domains and allows for expanded PAM recognition as well as increased specificity and lower off-target activity (Hu et al., 2018).
In 2020, the Kleinstiver lab reported the development of near-PAMless Cas9 variants. These variants, named SpG and SpRY, can target NGN PAMs and NRN and NYN PAMs, respectively (Walton et al., 2020).
Characterization of Cas9 from additional bacterial species
Many more Cas9 orthologs have been isolated, and our understanding of their PAMs is shown in the table below.
|
Cas9 species |
PAM sequence ’) |
|
Streptococcus pyogenes (Sp) |
3' NGG |
|
NGRRT or NGRRN |
|
|
NNNNGATT |
|
|
NNNNRYAC |
|
|
NNAGAAW |
|
|
NAAAAC |
|
|
PAM sequence may not be characterized |
Non-SpCas9's bind a variety of PAM sequences, which makes them useful when no suitable SpCas9 PAM sequence is present. Furthermore, non-SpCas9's may have other characteristics that make them more useful than SpCas9. For example, Cas9 from Staphylococcus aureus (SaCas9) is about 1 kilobase smaller than SpCas9, so it can be packaged into adeno-associated virus (AAV).
At 984 amino acids in length, Cas9 from Campylobacter jejuni (CjCas9) is even smaller than SaCas9 and is also compatible with AAV delivery. NmCas9, another small ortholog, displays lower off-target editing than wild-type SpCas9, even when targeting sites that are known to produce off-target editing with SpCas9 (Amrani et al., 2018). When choosing a Cas9, remember to check that your tracrRNA and crRNA (or synthetic gRNA) are derived from the same species.
Expanding the CRISPR toolbox
The isolation of novel CRISPR proteins has and will continue to dramatically increase the number of CRISPR applications. The first non-Cas9 CRISPR protein adapted for genome engineering was Cas12a (formerly Cpf1), a nuclease that generates double strand breaks in target genes resulting in the formation of “sticky ends” rather than the blunt ends created by Cas9. Cas12a displays lower off-target editing than SpCas9 (Kim et al., 2016). The Acidaminococcus sp. BV3L6 Cas12a, AsCpf1, uses a 5'-TTN PAM which makes it easier to target AT-rich genomes (Kleinstiver et al., 2016).
Type VI CRISPR systems, which target RNA, offer additional targeting flexibility. Cas13a (formerly C2c2) has been adapted for targeted RNA cleavage in mammalian cells. The REPAIR (RNA Editing for Programmable A to I Replacement) system fuses Cas13b to RNA deaminase ADAR2 to create a specific RNA editor. Cas13 enzymes are advantageous because they do not require a PAM, and RNA targeting is potentially reversible since there is no genomic edit. Some Cas13 enzymes require a single base protospacer flanking sequence (PFS) adjacent to the target, but many do not, showing the flexibility of this system.
As the number of CRISPR reagents continues to grow, so too will the number of reagents available through Addgene! If you're trying to decide which CRISPR tools are best for your experiment, check out our CRISPR guide or leave a comment below.
Note: Jennifer Tsang and Mary Gearing contributed to the update of this post.
References
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F (2017) RNA targeting with CRISPR–Cas13. Nature 550:280–284 . https://doi.org/10.1038/nature24049
- Find plasmids from this paper at Addgene.
Amrani N, Gao XD, Liu P, Edraki A, Mir A, Ibraheim R, Gupta A, Sasaki KE, Wu T, Donohoue PD, Settle AH, Lied AM, McGovern K, Fuller CK, Cameron P, Fazzio TG, Zhu LJ, Wolfe SA, Sontheimer EJ (2018) NmeCas9 is an intrinsically high-fidelity genome-editing platform. Genome Biol 19: . https://doi.org/10.1186/s13059-018-1591-1
Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F (2017) RNA editing with CRISPR-Cas13. Science 358:1019–1027 . https://doi.org/10.1126/science.aaq0180
- Find plasmids from this paper at Addgene.
Kim D, Kim J, Hur JK, Been KW, Yoon S, Kim J-S (2016) Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol 34:863–868 . https://doi.org/10.1038/nbt.3609
- Find plasmids from this publication at Addgene.
Kim E, Koo T, Park SW, Kim D, Kim K, Cho H-Y, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim J-S (2017) In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8: . https://doi.org/10.1038/ncomms14500
- Find plasmids from this publication at Addgene.
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, Aryee MJ, Joung JK (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481–485 . https://doi.org/10.1038/nature14592
- Find plasmids from this publication at Addgene.
Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK (2016) Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol 34:869–874 . https://doi.org/10.1038/nbt.3620
- Find plasmids from this publication at Addgene.
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556:57–63 . https://doi.org/10.1038/nature26155
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191 . https://doi.org/10.1038/nature14299
- Find plasmids from this publication at Addgene.
Walton RT, Christie KA, Whittaker MN, Kleinstiver BP (2020) Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368:290–296 . https://doi.org/10.1126/science.aba8853
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163:759–771 . https://doi.org/10.1016/j.cell.2015.09.038
- Find plasmids from this publication at Addgene.
Additional Resources on the Addgene Blog
- Read more about Cpf1
- Read more about SaCas9 and AAV
- Check out our CRISPR Featured Topic Page
Resources on Addgene.org
- Check out our CRISPR Guide
- Browse All CRISPR Plasmids
- Find Validated gRNAs
Topics: CRISPR, CRISPR 101, Cas Proteins







Leave a Comment