This post was written jointly by Addgenies Brook Pyhtila and Nicole Waxmonsky
 Resource sharing shortens the time needed to go from planning an experiment to performing one. At Addgene, over 120 labs have deposited CRISPR reagents, including many gRNA-containing plasmids (McDade et al, 2016). Many of the gRNAs contained within these plasmids have been used successfully in peer reviewed articles. If you’re targeting your favorite gene with CRISPR, using one of these validated gRNAs can save you the time that would be spent making and testing entirely new gRNA designs. You can now easily find many validated gRNAs in our newly curated Validated gRNA Target Sequence Table.
Resource sharing shortens the time needed to go from planning an experiment to performing one. At Addgene, over 120 labs have deposited CRISPR reagents, including many gRNA-containing plasmids (McDade et al, 2016). Many of the gRNAs contained within these plasmids have been used successfully in peer reviewed articles. If you’re targeting your favorite gene with CRISPR, using one of these validated gRNAs can save you the time that would be spent making and testing entirely new gRNA designs. You can now easily find many validated gRNAs in our newly curated Validated gRNA Target Sequence Table.
What are validated gRNAs?
Proper target selection and gRNA design are essential for a CRISPR experiment. The selection process is often time consuming and there is no guarantee that the selected sequence will yield a useful gRNA. Alternatively, you can select a target sequence which has already been shown to work in CRISPR experiments.
Addgene’s dedication to resource sharing has led us to develop a searchable and sortable datatable that contains validated gRNA sequences. Validated in this context refers to the fact that every gRNA listed in the table has been used effectively in a published article. This blog post will serve as a guide to use our validated gRNA Target Sequence Table.
Disclaimer: The efficacy of gRNAs is impacted by the target location, target sequence, and model system. Even if a gRNA has been demonstrated to work in your system, it is worth spending time sequencing the genomic target to determine whether any sequence variations (e.g. SNPs) exist. Additionally, consider testing multiple gRNAs targeted to the same locus to ensure their effects are specific to that locus.
The data in the validated gRNA table has been derived from scientists’ submissions, either their deposited gRNA plasmids or from submission of a spreadsheet containing the validated gRNA sequences alone. Sequence submission is welcomed for validated gRNAs that are not generated from a plasmid. If you, for example, prefer to deliver your CRISPR components as RNPs (ribonucleoproteins) and therefore usually transcribe your gRNAs in vitro, you can send us the sequences of your effective gRNAs, and we’ll add them to this list.
Access the spreadsheet for submitting your gRNA sequences here.
Features of the validated gRNA table
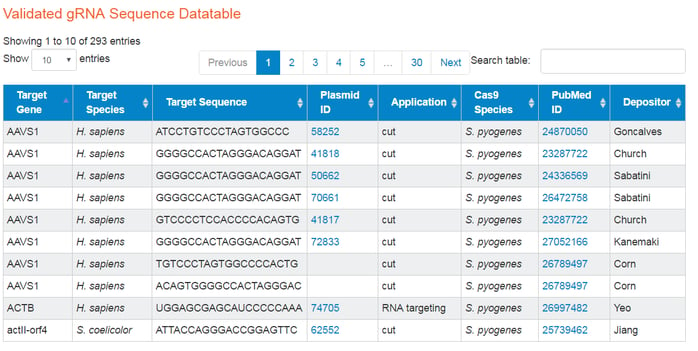 You can access the datatable here. Current table columns are target gene, species, sequence, Addgene plasmid ID (if we have one associated with the gRNA), application, cas9 species, PubMed ID, and depositor. The CRISPR applications we currently have data for are: cut, activate, interfere, visualize, nick, purify, tag, scaffold, CRISPR-display, and dCas9-FokI.
You can access the datatable here. Current table columns are target gene, species, sequence, Addgene plasmid ID (if we have one associated with the gRNA), application, cas9 species, PubMed ID, and depositor. The CRISPR applications we currently have data for are: cut, activate, interfere, visualize, nick, purify, tag, scaffold, CRISPR-display, and dCas9-FokI.
This table is both sortable and searchable, which can help exclude the data that you are not interested in. You can use the carats in the datatable headers to sort alphabetically or in ascending or descending order, in the case of numeric columns.
The data in any column are searchable and you can search by any part of the data. For example, entering ‘C. elegans’, or ‘elegans’ will filter the table to show C. elegans gRNA sequences. You can also filter by multiple keywords that are separated by a single space to get even more specific. For example, to show the gRNAs that were used for activation in a human system, search for ‘sapiens activate’ to filter the table and find exactly what you're looking for.
Search using the box highlighted in red above. When searching, it is worth trying alternate names, particularly for gene names.
If you find a sequence that will work for your experiment, it is recommended that you confirm the details and final experimental outcome in the original publication. As you develop and confirm new gRNAs, please consider submitting their sequences (and plasmids!) so that this shared resource can continue to grow.
This post was written jointly by Brook Pyhtila and Nicole Waxmonsky.
References
McDade J, Waxmonsky NC. Emerging CRISPR Technologies Available Through Resource Sharing. 2016. In press.
Additional Resources on the Addgene Blog
- Learn How to Design Your gRNA
- Use the CRISPR Software Matchmaker
- Browse Alll of Our CRISPR Posts
Additional Resources on Addgene.org
- Browse gRNA Design Tools
- Read Our CRISPR Guide Pages
- Find CRISPR Plasmids
Topics: Addgene News, Using Addgene's Website






Leave a Comment