
It’s time for another edition of “What’s New in CRISPR” - where we highlight a few of the newest CRISPR plasmids available at Addgene. If you want to read more content related to CRISPR technology, subscribe to our CRISPR blog content.
In this post we will cover:
- Miniature CRISPR-Cas systems
- SEND plasmids
- Prime editing improvements
Miniature CRISPR-Cas systems
One drawback to the commonly used S. pyogenes CRISPR/Cas9 system is that it is too large to be effectively packaged into AAV particles for delivery into mammalian cells, especially when fused to effector proteins for gene activation or gene editing. The more recently identified Cas12f systems from uncultivated archaea are much smaller and would be ideal for such AAV packaging, but their activity in mammalian systems has been in question. Three different labs have recently published work that provides a solution to this problem.
A miniature CRISPR-Cas system for mammalian use
Stanley Qi’s lab has developed a miniature Cas system (CasMINI) engineered from the uncultivated archaea Cas12f (Un1Cas12f1, or Cas14a1) system. The CasMINI system was developed by engineering the Cas12f scaffold gRNA and making strategic mutations in the DNA binding cavity of Cas12f. The CasMINI system is less than half the size of the Cas9 or Cas12a based systems, making it well suited for AAV packaging. Most importantly, this CasMINI system was shown to be effective for CRISPR activation, base editing, and genome editing in mammalian cells. The “V4” version (named CasMINI) had the highest activity for gene activation and base editing, whereas a different set of mutations (the “V3.1” version) works best for genome editing with an active nuclease.
- Read the paper in Molecular Cell
- Find the CasMINI plasmids
Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs
Yong-Sam Kim’s lab also created a miniature yet active Cas system starting with the same Un1Cas12f1 (Cas14). They noted that Cas12f1 had an extra-long gRNA for its small size. They were able to increase activity and decrease the gRNA size by intensively remodelling the gRNA scaffold through five rounds of gRNA engineering. They did not modify the Cas12f1 enzyme. The result is a highly efficient Cas system with very compact genome editors and a gRNA down-sized by almost 40%. The activity of Cas12f1 with these modified gRNAs is comparable to that of SpCas9. The group also confirmed that the Cas12f1 and modified gRNA were effective at genome editing when delivered in an AAV system, and demonstrated that the new system was able to activate expression by fusing dCas12f to VP64.
- Read the paper in Nature Biotechnology
- Find the Cas12f-GE plasmids
Programmed genome editing by a miniature CRISPR-Cas12f nuclease
Quanjiang Ji’s lab describes an effective miniature Cas system that utilizes a Cas12f from a different species, Acidibacillus sulfuroxidans, AsCas12f1. They found that this system, without any modifications, is effective in genome editing in both bacteria and human cells using various delivery methods, including plasmid, ribonucleoprotein, and AAV.
- Read the paper in Nature Chemical Biology
- Find the AsCas12f1 plasmids
Selective endogenous encapsidation for cellular delivery of RNAs
By Mike Lacy
The retroviral-like protein PEG10 specifically binds its own mRNA to facilitate its packaging into virus-like particles. Feng Zhang’s lab adapted the native PEG10 RNA sequences to instead drive packaging of cargo sequences in the capsid, in an approach called Selective Endogenous eNcapsidation for cellular Delivery (SEND). They demonstrate SEND as a modular system for packaging, secretion, and delivery of specific RNAs, such as Cre mRNA or Cas9 mRNA and sgRNA, in human and mouse cells. They further develop a version of SEND for gene transfer with pseudotyped viral particles, expanding options for CRISPR or other gene delivery.
- Read the paper in Science
- Find the SEND plasmids
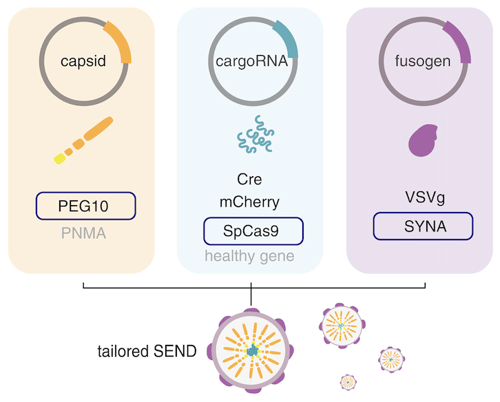 |
| Figure 1: SEND combines an endogenous Gag homolog, cargo RNA, and fusogen in a modular system for delivering gene editing tools into cells. Image adapted from Segel et al. 2021. |
Prime editing improvements
Several labs have recently shared some exciting plasmid tools to optimize prime editing systems.
Engineered pegRNAs improve prime editing efficiency
In the two years since David Liu’s group launched prime editing, they have continued to build upon knowledge of the biology of the system to optimize its outcomes. In their most recent work, the Liu Lab boosts the efficiency of prime editing to correct mutations in a variety of target genes and cell types. Their Nature Biotechnology paper showcases an improved design of the guide RNA that caps the 3’ end with a structured RNA motif. These engineered pegRNAs (epegRNAs) protect the RNA from degradation by cellular exonucleases and broadly improve editing efficiency.
- Read the paper in Nature Biotechnology
- Find the epegRNA plasmids
Enhanced prime editing systems by manipulating MMR machinery
In their Cell paper, the Liu Lab found that mismatch repair (MMR) interfered with the desired resolution of prime editing intermediates. Building upon the PE2 and PE3 systems of their earlier work, they developed a system in which the cellular MMR machinery is disrupted via a dominant negative mutation in MLH1 (PE4 = PE2+MLH1dn and PE5 = PE3+MLH1dn). Silent mutations added near the intended edit further weakened recognition by MMR machinery and improved editing efficiency. The lab also optimized the architecture and amino acid sequence of the prime editor, yielding PEmax, to work synergistically with epegRNAs and PE4 and PE5. These enhanced prime editing systems improve gene editing performance at therapeutically relevant gene targets and expand the application to the possible treatment of genetic disease.
- Read the paper in Cell
- Find PE4, PE5, and PEmax plasmids
Precise genomic deletions
In PRIME-del, developed by Jay Shendure’s Lab, paired pegRNAs work with the PE2 enzyme from the Liu lab to target opposite DNA strands and specify a large intervening deletion. Not only can this method precisely program large genomic deletions of up to 10kb but it is also capable of coding concurrent small insertions to allow in-frame deletions and introduce epitope tags and RNA transcription start sites. The lab further demonstrated that PRIME-del can be “multiplexed” using pooled paired pegRNAs.
- Read the paper in Nature Biotechnology
- Find the PRIME-del plasmids
Deletion and replacement of long genomic sequences
The PEDAR strategy engineered by Wen Xue’s lab also uses paired pegRNAs targeting complementary DNA strands to program large genomic deletions with or without small insertions. But, instead, PEDAR couples this deletion process to a prime editing enzyme with a fully active Cas9 enzyme. PE-Cas9 comprises an active Cas9 fused to the reverse transcriptase (RT), which induces two double-strand breaks and deletes the intervening sequence. As with other prime editing tools, the desired edits are templated by the 3′ extension of the pegRNAs. PEDAR is capable of correcting larger genomic rearrangements as demonstrated in a mouse model of tyrosinemia, where a pathogenic insertion within the FAH gene was precisely corrected, restoring expression in the liver.
- Read the paper in Nature Biotechnology
- Find the PEDAR plasmids
Additional resources on the Addgene blog
- Download the CRISPR 101 eBook
- Find our CRISPR 101 blog posts
- Read our CRISPR cheat sheet
Resources on Addgene.org
- Read our CRISPR guide
- Find CRISPR plasmids by function
- Find recently deposited plasmids
Topics: CRISPR, Other CRISPR Tools
Topics: CRISPR






Leave a Comment