Direct reprogramming describes the process where differentiated cells are turned into a cell type of choice while bypassing the intermediate pluripotent state. Though a valuable tool for regenerative medicine, direct reprogramming is an inefficient process, with the majority of cells failing to develop the desired identity.
The development of single cell technologies, such as single cell RNA-seq (scRNA-seq), now allows scientists to identify the gene expression patterns unique to the few cells that successfully reprogram. Unfortunately scRNA-seq only captures the gene expression of cells at a single time point, making it difficult to investigate how early expression patterns influence reprogramming outcomes.CellTag from the Morris Lab at Washington University in St. Louis provides a combinatorial indexing strategy that simultaneously tracks clonal history and cell identity at the single-cell level, thus linking ancestral gene expression patterns with reprogramming outcomes (Biddy et al., 2018).
Tracking lineage and cell identity at the single-cell level with CellTag
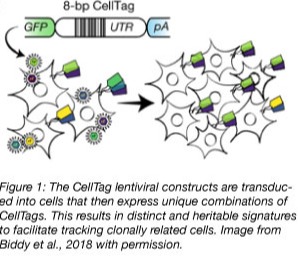 CellTagging is a lentiviral based tagging method that labels cells with heritable barcodes. Barcodes consist of an 8bp variable region and a 6 bp fixed region. The variable region acts as a unique identifier for individual cells. Each cell is labeled with 3-4 tags to increase the diversity of the CellTag library. The fixed region acts as a library-specific identifier and allows cells to be tagged with different CellTag libraries at multiple timepoints during reprogramming. These library-specific identifiers act like a timestamp and help demultiplex the scRNA-seq data and build multi-generational cell lineage trees. CellTags were detected over a 10-week period. Their expression decreased over time but they were not silenced.
CellTagging is a lentiviral based tagging method that labels cells with heritable barcodes. Barcodes consist of an 8bp variable region and a 6 bp fixed region. The variable region acts as a unique identifier for individual cells. Each cell is labeled with 3-4 tags to increase the diversity of the CellTag library. The fixed region acts as a library-specific identifier and allows cells to be tagged with different CellTag libraries at multiple timepoints during reprogramming. These library-specific identifiers act like a timestamp and help demultiplex the scRNA-seq data and build multi-generational cell lineage trees. CellTags were detected over a 10-week period. Their expression decreased over time but they were not silenced.
Applications of CellTag
The Morris Lab developed CellTag to track the direct reprogramming of mouse embryonic fibroblasts (MEFs) into induced endoderm progenitors (iEPs), a self-renewing cell type that can differentiate into hepatic or intestinal cell types. MEFs were reprogrammed into iEPs by retroviral expression of transcription factors Foxa1 and Hnf4α, a process that like other reprogramming protocols is inefficient.
Cells were sequentially tagged with different CellTag libraries at three time points during the 28 day study: prior to reprogramming (CellTagMEF), at the end of the three-day transcription factor reprogramming treatment (CellTagD3), and 13 days after the start of reprogramming (CellTagD13), which is when the iEP phenotype typically emerges. Every three days, a sample of cells was collected for scRNA-seq analysis and the remaining cells were maintained in culture. iEP cell physiology and reprogramming efficiency was not affected by CellTagging.
By constructing lineage trees and comparing them with the gene expression changes of more than 100,000 cells undergoing iEP conversion, the Morris Lab found that cells took one of two trajectories: a route towards successful iEP conversion or a ‘dead-end' path where cells were partially reprogrammed and re-expressed fibroblast genes. Most cells ended up taking the ‘dead-end’ path. The CellTags revealed that cells with common ancestors tended to have similar reprogramming outcomes. For example, cells with shared CellTagD13 tags often had similar gene expression pattern changes and reprogramming outcomes, suggesting that outcomes are already determined by at least day 13. However, distinct transcriptional states were seen as early as day 6. All together these results suggest that iEP reprogramming is not stochastic, but rather there’s a particular cell state that’s permissive to reprogramming.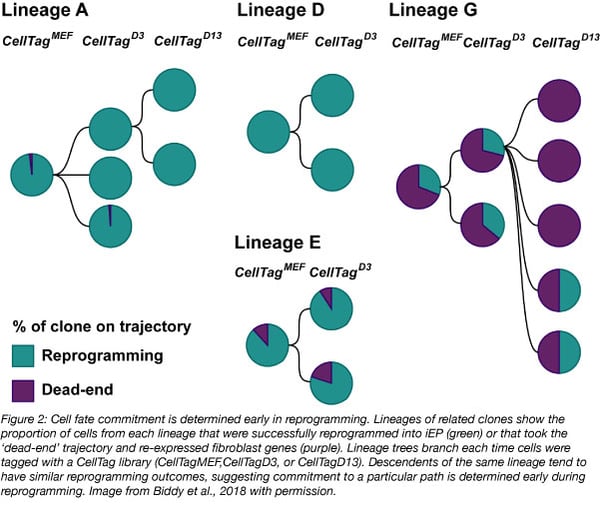 In this lineage tracing experiment, the Morris lab identified a novel gene, Mettl7a1, whose expression helped coax cells towards successful iEP reprogramming. MettI7a1 encodes an uncharacterized putative methyltransferase. Adding Mettl7a1 to the standard Foxa1-Hnf4α retroviral overexpression cocktail yielded a three-fold increase of fully reprogrammed cells based on single-cell gene expression patterns.
In this lineage tracing experiment, the Morris lab identified a novel gene, Mettl7a1, whose expression helped coax cells towards successful iEP reprogramming. MettI7a1 encodes an uncharacterized putative methyltransferase. Adding Mettl7a1 to the standard Foxa1-Hnf4α retroviral overexpression cocktail yielded a three-fold increase of fully reprogrammed cells based on single-cell gene expression patterns.
The lab has also used CellTags for tagging two cell populations in a competitive transplant assay in the mouse intestine, demonstrating its utility in long-term tracking in vivo (Guo et al., 2019).
Key takeaways of CellTag
CellTag is a combinatory cell-indexing tool that combines scRNA-seq and lineage tracking to track the dynamics of gene expression and clonal history associated with changes in cell identity. The Morris Lab used CellTagging to uncover two distinct trajectories of MEFs undergoing iEP conversion, but CellTag could be used to study other biological processes where cells are undergoing identity changes, such as the differentiation of induced pluripotent stem cells or the clonal evolution of cancer.
If you’re looking to use CellTag, you can find the CellTag libraries at Addgene. The Morris Lab's website also has several useful resources including a link to a detailed CellTag protocol on protocols.io, code and analysis tutorials on GitHub, and a CellTag lineage data visualization tool.
 Check out the CellTag webpage!
Check out the CellTag webpage!
References
Biddy, Brent A., et al. "Single-cell mapping of lineage and identity in direct reprogramming." Nature 564.7735 (2018): 219. PubMed PMID: 30518857.
Guo, Chuner, et al. "CellTag Indexing: genetic barcode-based sample multiplexing for single-cell genomics." bioRxiv (2019): 335547.
Additional resources on the Addgene blog
- Learn more about induced pluripotent stems cells (iPSCS) and reprogramming and direct reprogramming
- Explore different methods for delivery of reprogramming factors for generating iPSCs
- Learn how fluorescent proteins help researchers study single cell dynamics in model organisms
Resources on Addgene.org
- Visit our stem cell page
- Find plasmids for reprogramming and differentiation
- Check out our lentivirus guide
- Sign up to receive our Viral Vectors 101 eBook
Topics: Viral Vectors, Cell Tracing





Leave a Comment