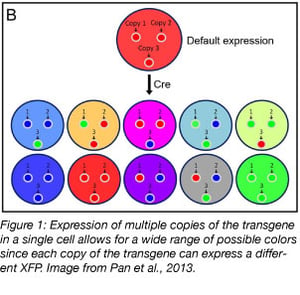
 Stochastic multicolor labeling is a popular technique in neuroscience and developmental biology. This type of cell labeling technique involves the introduction of a transgene construct containing fluorescent proteins (XFP) of different colors to label an organ or entire organism. Because each cell can have multiple copies of the transgene that will recombine independently, cells may acquire one of a variety of colors when a combination of XFP are expressed. Each cell remains the same color for its entire lifetime and daughter cells retain the same color, allowing for the fate mapping of cell populations over time. The ability to track single cell dynamics at the organism level has been made possible by tools that allow cells to become persistently fluorescent during development. Stochastic multicolor labeling systems, many based on Brainbow, now exist for a variety of species, cell types, and research applications.
Stochastic multicolor labeling is a popular technique in neuroscience and developmental biology. This type of cell labeling technique involves the introduction of a transgene construct containing fluorescent proteins (XFP) of different colors to label an organ or entire organism. Because each cell can have multiple copies of the transgene that will recombine independently, cells may acquire one of a variety of colors when a combination of XFP are expressed. Each cell remains the same color for its entire lifetime and daughter cells retain the same color, allowing for the fate mapping of cell populations over time. The ability to track single cell dynamics at the organism level has been made possible by tools that allow cells to become persistently fluorescent during development. Stochastic multicolor labeling systems, many based on Brainbow, now exist for a variety of species, cell types, and research applications.
Labeling neurons with Brainbow
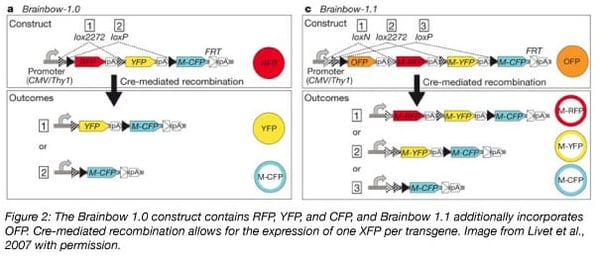
The Lichtman group developed the original iteration of this technology in 2007. Used to label neurons in the cerebellum of mice, the team appropriately named their technology Brainbow. Brainbow 1.0 used the fluorophores RFP (red), YFP (yellow), and membrane-tethered CFP (cyan). Brainbow 1.1 adds OFP (orange) (Livet et al., 2007).
Brainbow uses a Cre/lox system to label neurons. Genes encoding the fluorescent proteins are organized in tandem with a single promoter, but only the gene directly following the promoter is expressed. Cre-mediated deletion of one or several of these genes permits the expression of genes farther downstream in the construct. As RFP and OFP are directly downstream of the promoter in Brainbow 1.0 and 1.1, respectively, cells will fluoresce either red or orange until recombination occurs.
Following the development of Brainbow 1.0, several more refined versions have been developed, which differ in the number, stability, and brightness of the fluorescent proteins used in the system.
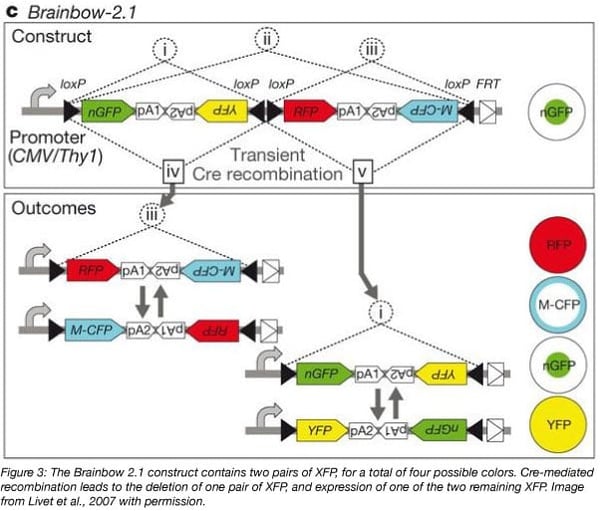
One such system is Brainbow 2.0, which uses Cre to trigger the inversion of two genes aligned head-to-head. Another variation, Brainbow 2.1, uses Cre to trigger the inversion of two tandem sets of genes aligned head-to-head and the deletion of one of the gene pairs. Brainbow 2.0 therefore acts as a switch between the default RFP and CFP, while Brainbow 2.1 switches from the default GFP (green) to RFP, CFP, or YFP fluorescence. In Brainbow 2.1, GFP is tagged with a nuclear localization sequence, allowing it to localize to a different area of the cell than CFP and YFP.
The most recent derivation of Brainbow, Brainbow 3.0 (Cai et al., 2013), incorporates the highly photostable fluorophores mOrange2, EGFP, and mKate2 (red) to solve the problematically low fluorescent intensity of previous versions of Brainbow. Developed from Brainbow 3.0 and using the same fluorescent markers, Brainbow 3.1 and 3.2 (described in the same publication) are unique because by default, they are not fluorescent. Instead, they incorporate a truncated, non-fluorescent form of YFP as the default state of the transgene such that recombination is necessary to excise the non-fluorescent YFP and activate fluorescence of the downstream OFP, GFP, or RFP. Cells that express the construct but do not undergo recombination can be detected by immunostaining of the truncated YFP.
In Thy1-Brainbow mice, some neurons expressed as many as 16 copies of the injected transgene, each of which could recombine to activate the expression of a different fluorescent protein. Therefore, each cell could express a unique combination of all of the fluorescent proteins from the injected construct. The fluorescent protein expressed in a single cell maintains its color over the entire length of the cell, even axons of up to 100 microns in length.
Some challenges of the Brainbow system are that it does not use ubiquitous promoters, meaning that not all cell types can be labeled, and that while multiple copies of the transgene can integrate into cells, the expression level of these XFP is not strong enough for all applications. To resolve this, similar systems have been developed for tracking various cell types and organisms and for improving the level of detail possible for stochastic multicolor labeling of the brain.
Labeling Drosophila neurons with dBrainbow and Flybow
Based on the Brainbow 1.0 system, Drosophila Brainbow (dBrainbow; Hampel et al., 2011) labels Drosophila neurons at the neuroblast stage, meaning that neurons of the same lineage will express the same XFP transgene in an adult fly. As a stop cassette is located directly upstream of the fluorescent protein genes, Cre recombination is required before fluorescence can occur. Cre recombinase can be used alongside common, existing tools for fly research that use Flp recombination. In addition to the XFP in the construct, dBrainbow includes small epitopes fused to each XFP. Secondary antibodies that target these epitopes and fluoresce in the same color as the fused XFP can be used to amplify the signal. Imaging endogenous fluorescence in live animals only permits visualization of two colors.
Similarly, Flybow adapts Brainbow 2.0 for use in Drosophila melanogaster and is under the control of Flp recombinase. The XFPs in Flybow are membrane-tethered, making this system useful for visualization of cellular extensions, such as neurons in the fly visual system, which has been difficult to study previously (Hadjieconomou et al., 2011).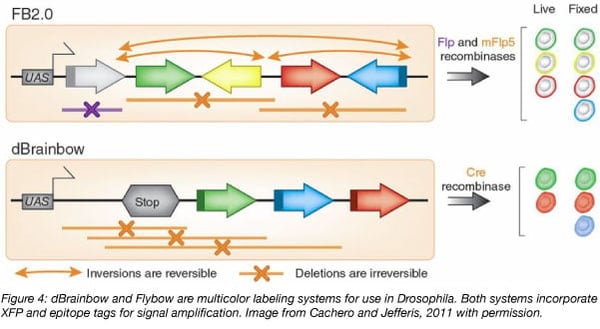
Cell tracing in zebrafish with Zebrabow
Before the generation of Zebrabow (Pan et al., 2011), Brainbow was also used in zebrafish by injecting a CMV-Brainbow construct at the embryonic stage. This technique could be used to target Brainbow to neurons by placing Brainbow downstream of a neural-specific promoter, or the entire fish could be labeled by injecting Brainbow at the single-cell stage. Zebrabow, or “zebrafish brainbow,” is a collection of transgenic zebrafish lines that allow for multicolor labeling of an entire organism or a specific tissue. XFPs are introduced using a plasmid construct called Zebrabow-GateDest containing an RFP, CFP, and YFP downstream of a ubiquitous promoter. Zebrabow is an extremely versatile technology as recombination can be induced in developing fish via Cre protein microinjection or crossing of Zebrabow animals to an inducible Cre line. The Schier group developed Zebrabow as a system that takes advantage of the multicolor labeling possible from Brainbow.
Tracing clonal cell populations using the Confetti mouse
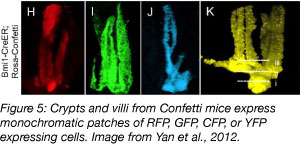 For mammalian systems, the Clevers lab developed the Confetti mouse (Snippert et al., 2010), a reporter mouse that can be used to track any cell type when crossed with mice expressing the appropriate Cre allele. The Confetti mouse contains the Brainbow 2.1 construct in the Rosa26 locus, under a loxP “roadblock” so that XFP are only expressed following a recombination event. Tissue-specific Cre recombination in this mouse generates cells that randomly express either nuclear GFP, membrane CFP, cytoplasmic YFP, or cytoplasmic RFP in a specific cell type.
For mammalian systems, the Clevers lab developed the Confetti mouse (Snippert et al., 2010), a reporter mouse that can be used to track any cell type when crossed with mice expressing the appropriate Cre allele. The Confetti mouse contains the Brainbow 2.1 construct in the Rosa26 locus, under a loxP “roadblock” so that XFP are only expressed following a recombination event. Tissue-specific Cre recombination in this mouse generates cells that randomly express either nuclear GFP, membrane CFP, cytoplasmic YFP, or cytoplasmic RFP in a specific cell type.
The Confetti mouse was first used to describe the self-renewal of cells in the intestinal crypt, and since then has become a powerful tool for understanding cell proliferation.
Inducible XFP expression in the Ubow mouse
Another murine reporter animal, the Ubow mouse (Ghigo et al., 2013), was developed from the Brainbow 1.0-L construct, a variant of Brainbow 1.0 that contains RFP, CFP, and YFP. After crossing onto a tamoxifen-inducible ubiquitin background, each cell in the mouse that expresses Cre undergoes exactly one recombination event, leading to each copy of the transgene expressing either YFP or CFP, and cells that do not express Cre fluorescing with the default RFP.
The Ubow construct was then used to develop a system for tracking the fate of Langerhans cells. In the Ubow mouse, cells expressing YFP, CFP, and RFP are not present at the same frequencies. The small population size of CFP++ cells permitted researchers to longitudinally map the turnover of Langerhans cells in the dermis and identify that Langerhans cells are replenished through the proliferation of specific, and not all, Langerhans cells, to create foci of proliferating cells.
Tetbow optimizes Brainbow for current technologies
Tetbow (Sakaguchi et al., 2018) overcame the low expression of XFP of Brainbow by incorporating a Tet-Off system to boost XFP expression. The increased brightness of Tetbow allows this system to be used to trace neurons over millimeter-long distances across multiple fields of view, whereas previous Brainbow technologies were useful for discriminating neurons within only one field of view. Tetbow can be introduced in vivo either by electroporation of plasmids, or using AAV vectors.
The Imai lab also developed a version of Tetbow that incorporates chemical tags instead of XFP to optimize it for tissue clearing, a method that makes tissues, such as the brain, transparent so that the 3D architecture of a fluorescent neuron is visible. Chemical tags are more resistant to tissue clearing methods and can be labeled by synthetic fluorophores, which are more stable than fluorescent proteins. For more details, check out this blog post from the researchers who developed Tetbow.
Conclusion
Stochastic multicolor cell labeling systems allow single cell dynamics in a variety of organisms to be observed in more detail than ever before. As scientists continue to develop new iterations of these technologies, expect to see systems that use newer, more photostable fluorophores and that can be combined with new imaging techniques.
References
Cai, Dawen, et al. "Improved tools for the Brainbow toolbox." Nature methods 10.6 (2013): 540. PubMed PMID: 23817127. PubMed Central PMCID: PMC3713494.
Ghigo, Clément, et al. "Multicolor fate mapping of Langerhans cell homeostasis." Journal of Experimental Medicine 210.9 (2013): 1657-1664. PubMed PMID: 23940255. PubMed Central PMCID: PMC3754858.
Hadjieconomou, Dafni, et al. "Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster." Nature methods 8.3 (2011): 260. PubMed PMID: 21297619.
Köhler, Corinna, et al. "Mouse cutaneous melanoma induced by mutant BRaf arises from expansion and dedifferentiation of mature pigmented melanocytes." Cell stem cell 21.5 (2017): 679-693. PubMed PMID: 29033351.
Livet, Jean, et al. "Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system." Nature 450.7166 (2007): 56. PubMed PMID: 17972876.
Pan, Y. Albert, et al. "Multicolor Brainbow imaging in zebrafish." Cold Spring Harbor Protocols 2011.1 (2011): pdb-prot5546.
Richardson, Alexander, et al. "Visualizing the Fate of Transplanted K14-Confetti Corneal Epithelia in a Mouse Model of Limbal Stem Cell Deficiency." Investigative ophthalmology & visual science 59.3 (2018): 1630-1640.
Sakaguchi, Richi, Marcus N. Leiwe, and Takeshi Imai. "Bright multicolor labeling of neuronal circuits with fluorescent proteins and chemical tags." eLife 7 (2018): e40350. PubMed PMID: 30454553. PubMed Central PMCID: PMC6245733.
Snippert, Hugo J., et al. "Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells." Cell 143.1 (2010): 134-144. PubMed PMID: 21205846. PubMed Central PMCID: PMC3082469.
Tarlow, Branden D., Milton J. Finegold, and Markus Grompe. "Clonal tracing of Sox9+ liver progenitors in mouse oval cell injury." Hepatology 60.1 (2014): 278-289. PubMed PMID: 24700457. PubMed Central PMCID: PMC4077948.
Yan, Kelley S., et al. "The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations." Proceedings of the National Academy of Sciences 109.2 (2012): 466-471. PubMed PMID: 22190486. PubMed Central PMCID: PMC3258636.
Additional resources on the Addgene blog
- Read an introduction to live cell imaging
- Learn more about the origin of the Brainbow system
- Visit our fluorescent proteins topic page
Resources on Addgene.org
- Browse Addgene’s fluorescent protein collection
- Learn about fluorescent tools for in vivo imaging
Topics: Fluorescent Proteins, Cell Tracing, Neuroscience






Leave a Comment