Why study neural connectivity?
One of the early lessons many of us learned in biology is that the body’s architecture and plumbing are important. We started with learning the head is connected to the neck. Shortly after, we learned about organs and the jobs they perform. This became foundational later on when we studied biological processes, like how our stem cells are housed in specific locations and give rise to progenitors during growth and development or that blood flows through the heart and lungs and oxygenates the body. However, in neuroscience, this architecture is frequently still an open question. The connections between neurons are what define how the brain operates, and thus, are a major part of the answer to many biological questions about the brain. To address this, molecular tools to map neuronal connectivity are widely used in neuroscience. In this post, I’ll describe how rabies virus (RABV) can be used in the brain to visualize how neurons are connected.
 Viral Vectors as Neuronal Tracers
Viral Vectors as Neuronal Tracers
Due to their ability to efficiently enter cells and deliver a variety of cargoes, genetically engineered neurotropic viruses emerged as powerful tools for visualizing neural connectivity (Nassi et al. 2015). The recombinant viruses typically used in neuroscience research (e.g., AAV) are useful for labeling the individual neurons they infect. However, they generally are not transmitted to adjacent neurons via synapses (i.e., transmitted transsynaptically). This is because the recombinant forms of these viruses typically used in research aren’t able to replicate, and for a virus to cross a synapse from one cell to another, it must replicate.
Modified Rabies Virus: Converting a transsynaptic neuronal tracer into a monosynaptic (first order) neuronal tracer
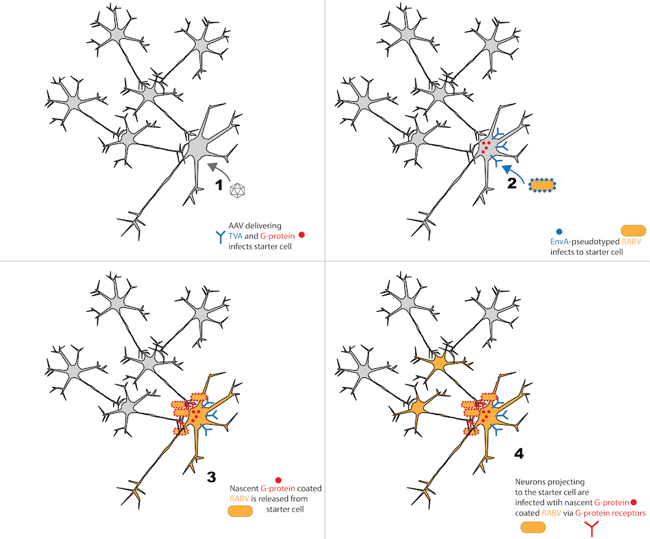
For RABV to crawl through the whole brain, it first infects one cell (the “starter cell”). It then replicates within that cell, and nascent RABV cross synapses to retrogradely infect neurons that project to the starter cell. Once this first order infection occurs, subsequent cycles of viral replication and retrograde transmission occur, spreading the virus through higher and higher order neural connections. While this transsynaptic spread is useful for neuronal tracing, it has two practical limitations. First, these high levels of viral replication are toxic. Second, infecting subsequently higher and higher order neurons doesn’t reveal network connections in a useful way, but rather just labels the whole brain.
To achieve labeling of only first-order connections to the starter cell, RABV was modified to only infect user-defined starter cell types, as well as to only spread monosynaptically from these cells.
These modifications take advantage of the fact that RABV is an enveloped virus. In general, viral envelopes dictate infection of target cells through their interactions with target cell surface receptors (Callaway 2008). Based on this, cell type specificity of the starter cell can be achieved by using EnvA-pseudotyped RABV, and only expressing the EnvA receptor (TVA) in user-defined cells. For example, a researcher can deliver Cre-dependent TVA to Cre-expressing neurons in a transgenic mouse line. By doing this, RABV expressing the viral protein EnvA on its surface will only infect Cre-positive, TVA-expressing cells (Osakada et al., 2013). Ultimately, this allows RABV to selectively infect a genetically defined cell population. While the EnvA genetic sequences are supplied during production of EnvA-pseudotyped RABV, these sequences are not copied into viral genome, so viral progeny will not necessarily express EnvA unless it is supplied in trans.
Monosynaptic RABV spread can be achieved by modifying the wild-type RABV genome, which encodes a single envelope protein called rabies glycoprotein (G). G-protein-coated RABV infects cells via the G receptor. In neurons, the G receptor is restricted to presynaptic nerve terminals (Lafon 2005). Therefore, once RABV is released from an infected cell, it is only taken up by the presynaptic nerve terminals of adjacent neurons (i.e., it is taken up by projection neurons). Research into these phenomenon has shown that the RABV G protein is necessary and sufficient for retrograde transmission (Etessami et al., 2000, Wickersham et al., 2007). Thus, RABV transsynaptic spread can be controlled by selectively supplying the G protein in trans only to starter cells (infected with G-deleted RABV). In this case, G-deleted RABV will infect the starter cell. RABV replication will then occur and the G protein (supplied in trans) will be incorporated into the viral envelope (but not the viral genome). Nascent G protein-coated RABV is then released from the starter cell and transmits retrogradely to first order neurons, via entry mediated by the G protein/G-protein receptor. Once in first order neurons, the G-deleted RABV can divide and replicate (since these processes are mediated by other viral proteins), but the newly released RABV cannot infect any subsequent cells due to lack of G protein on the viral envelope (Wickersham et al., 2007).
Summary and RABV applications
Use of G deleted RABV in conjunction with restricted TVA and G protein expression is a powerful development in neural tracing. Using these technologies in combination with molecular tools (e.g., optogenetic or chemogenetic receptors) enables complex control of specifically defined neuronal populations.
In one study, RABV monosynaptic tracing was used to examine how adult-generated neurons integrated into the brain over time (Deshpande et al., 2013). The study found that at early time points, most of the projections to these new neurons came from nearby neurons. It wasn’t until later time points that longer-range connections were seen. This study addressed the question of how newly generated neurons integrate into the complex pre-existing circuit of the brain.
Another study used RABV monosynaptic tracing to identify inputs to two major dopaminergic cell populations in two specific brain regions (Watabe-Uchida et al., 2012). They could also quantify the strength of the inputs by counting the monosynaptic inputs. This study revealed that these neurons received more inputs than was previously thought. The differences in the inputs to each of the two populations could be used to explain the differences in the functions of these two neuronal populations. Overall, this study laid the foundation for how different anatomical regions of the brain receive inputs and compute outputs.
In general, RABV monosynaptic tracing is a powerful tool that can be used to reveal brain circuitry, and continue to contribute answers to the complex questions of how the brain operates.
References
1. Callaway EM. Transneuronal Circuit Tracing with Neurotropic Viruses. Curr Opin Neurobiol. 2008 Dec; 18(6): 617–623. Pubmed PMID 19349161.
2. Deshpande A, Bergami M, Ghanem A, Conzelmann KK, Lepier A, Götz M, Berninger B. Retrograde monosynaptic tracing reveals the temporal evolution of inputs onto new neurons in the adult dentate gyrus and olfactory bulb. Proc Natl Acad Sci U S A. 2013 Mar 19;110(12):E1152-61 Pubmed PMID 23487772.
3. Etessami R, Conzelmann KK, Fadai-Ghotbi B, Natelson B, Tsiang H, Ceccaldi PE. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J Gen Virol. 2000 Sep;81(Pt 9):2147-53. Pubmed PMID 10950970.
4. Lafon M. Rabies virus receptors. J Neurovirol. 2005;11:82–87. PubMed PMID 15804965.
5. Nassi JJ, Cepko, CL, Born RT, Beier KT. Neuroanatomy goes viral! Front Neuroanat. 2015 Jul 1;9:80. Pubmed PMID 26190977.
6. Osakada F, Callaway EM. Design and generation of recombinant rabies virus vectors. Nat Protoc. 2013;8:1583–1601. doi: 10.1038/nprot.2013.094. Pubmed PMID 23887178.
7. Watabe-Uchida M1, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012 Jun 7;74(5):858-73. Pubmed PMID 22681690.
8, Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Retrograde neuronal tracing with a deletion-mutant rabies virus. Pubmed PMID 17179932.
Additional Resources on the Addgene Blog
- AAVs CREATed for Gene Delivery to the CNS and PNS
- Advanced Uses of Cre-lox and Flp-FRT - A Neuroscientist's View
- Control Gene Expression with FLEx Vectors
Resources on Addgene.org
Topics: Viral Vectors, Cell Tracing







Leave a Comment