This post was contributed by guest blogger Katrin Michel.
Cre-lox is an incredibly popular and powerful site specific recombinase (SSR) system, but it only gives you a single level of control without modification - either Cre is there or it’s not. Cre-mediated possibilities for site specific (and often cell type specific) control of DNA recombination and gene expression can be advanced by the coordinated use of fellow SSR system FLP-FRT. In addition, a variety of means to spatiotemporally control FLP and Cre expression have been developed. Read on to learn more about FLP-FRT, Cre-lox, and how combinations of FLP and Cre enable additional levels of genetic control.
What is FLP FRT and how does it work?
FLP recombinase, which is named after its ability to invert or ‘flip DNA’, is derived from the yeast S. cerevisiae (Gronostajski and Sadowski 1985). The FLP system works in a similar fashion to the Cre system. The FLP recombinase recognizes FRT sites and initiates site directed gene recombination between those sites. FRT sites and loxP sites differ at the nucleotide level but share an overall structure of 13 base pair palindromic repeats separated by an 8 bp asymmetric core. Recombination between these sites can lead to excision, inversion, and translocation of DNA in a similar fashion as between loxP sites (For details see Cre-lox blog post).
Optimizing FLP FRT through smart engineering
Different FLP versions – making FLP efficient
The first version of FLP discovered has a temperature optimum of 30 °C and is therefore inefficient in mammalian cells (usually grown at 37 °C). Smart molecular evolution led to the identification of FLPe which has a temperature optimum of 37 °C (Buchholz, Angrand, and Stewart 1998). FLPe’s performance was further improved, by adding a nuclear localization sequence (SV40 Large T nuclear localization sequence) and expressing it from a CAGGs promoter. However FLPe is still less efficient than Cre recombinase. FLPe codon optimization resulted in FLPO, which shows similar efficiency to Cre (Kranz et al. 2010).
These differences in recombinase efficiency are important to keep in mind when deciding whether to use the Cre-lox or one of the FLP-FRT systems in your experiments.
Mutated FRT and loxP sites in Flex Vectors – inversion only once
All recombination events mediated by FLP or Cre are reversible. Whereas the excision of a piece of DNA flanked by loxP/FRT sites is favored over its reintroduction, inversion and re-inversion happen at the same probability. Continuous inversion of the DNA sequence of interest can lead to poor expression of genes within that sequence and should therefore be prevented.
The loxP and FRT target sites have been engineered to avoid this re-inversion issue. It became possible to design FLEx vectors (Flip-excision vectors) that only allow one inversion event when it was discovered that it is not the exact sequence of the asymmetric target site core but its 8 bp length that is critical for Cre and FLP function. Because recombination can only occur between target sites of the same sequence in FLEx vectors, the DNA sequence that should be inverted is flanked by two pairs of target sites that differ in their sequence. Recombination between the two target sites with identical sequence leads to inversion of the flanked DNA sequence and the interior target sites. In a second step, the two compatible target sites are cleaved out, leaving behind a vector with the inverted DNA flanked by non-compatible target sites (Schnütgen et al. 2003).
Flex vectors revolutionize neuroscience
Manipulation of specific cell types in neuronal networks
Double inverted vectors expressing light inducible ion channels (plasmid # 20298 for example) have revolutionized Neuroscience. They are the only means to target and manipulate specific cell types (populations) within neuronal networks in living animals. To accomplish such cell specific manipulations in vivo, FLEx vectors expressing the light activatable ion channel under the control of Cre recombinase are used in combination with mouse lines that express Cre only in the cell type of interest (Atasoy et al. 2008; Cardin et al. 2009).
Ligand regulated recombinase - keeping Cre out until needed
But what can be done when the recombination should only occur at a specific time point? To allow temporal control, ligand regulated Cre and FLPe recombinase have been developed. A common means to achieve ligand control is to fuse a mutant estrogen receptor (ER) ligand binding domain (LDB) to the C-terminus of FLP or Cre (pCAG-CreERT2 #14797). These Cre/ FLP versions are retained in the cytoplasm until estrogen receptor ligand Tamoxifen is added. Tamoxifen then binds to the ER-LDB domains of the fusions allowing Cre/FLP translocate to the nucleus where they exert their recombinase activity (Logie and Stewart 1995; Metzger et al. 1995).
Visualizing single neurons in the living brain
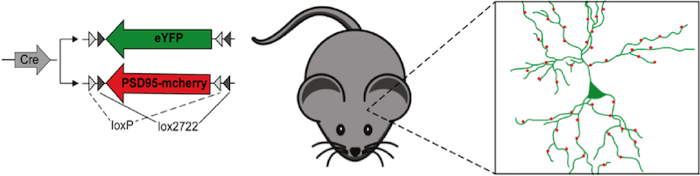 |
|
Figure 1: Plasmid mix to label neuronal morphology (eYFP) and the synaptic protein PSD95 (PSD95-mcherry) under the control of Cre recombinase. After in-utero electroporation, this plasmid mix results in sparse labeling of cortical neurons and their synapses. |
How can you label a single neuron and its synapses in the living brain? Cells of a specific brain area can be transfected by in utero electroporation. To achieve sparse labeling of individual neurons one could reduce the plasmids amounts to reduce transfection efficiency. However, if the cellular structure of a neuron and its synapse should be visualized at the same time from two different plasmids this approach becomes problematic as it reduces the probability that neurons are transfected with both vectors. Flex vectors are the solution.
Flex vectors expressing fluorescent cytoplasmic markers (e.g. eYFP) and synaptic proteins (e.g. postsynaptic PSD95-mcherry) are used in high concentrations in combination with low concentrations of vectors expressing the required recombinase. The high concentration of FLEx vectors guarantees that most cells are transfected with all labeling plasmids and the low concentration of recombinase restricts their expression to individual cells. With this technique, a Neuroscientist can see the full dendritic arbor of a single cell with all its synapses and how it rearranges these synapses in the living brain over time (Villa et al. 2016).
Combing FLP and Cre for additional control
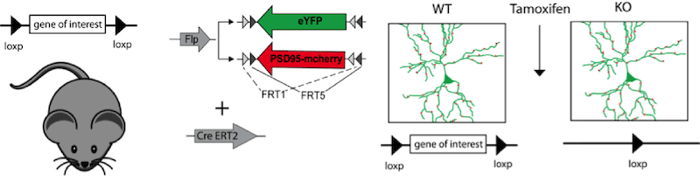 |
|
Figure 2: Expression of a morphological marker (eYFP) and synaptic marker (PSD95-mcherry), under the control of Flp recombinase together with inducible Cre recombinase (CreERT2), visualizes individual neurons and their synapses in a conditional knockout mouse. Tamoxifen injection induces Cre-ERT2 recombinase, leading to recombination of the loxP sites and thereby to the gene knockout. In this experiment it is possible to compare neuronal morphology and synaptic connectivity before and after the gene knockout in the same mouse. |
Scientists always strive for more. In order to analyze the function of individual proteins in neuronal circuits or synaptic remodeling, researchers combine the Cre and FLP recombinases to control both knockout of a target gene and expression of a separate gene. In these mouse lines, gene knockout can be induced by Cre recombinase expression while proteins needed for later experiments (cellular tracking for instance) can be controlled with FLP-FRT. For further temporal control, Tamoxifen inducible Cre versions can be used in these experiments. This elegant combination of Cre, FLP, and inducible systems gives a researcher exquisite control over expression of multiple genes in specific cells at specific times.
Many thanks to our guest blogger, Katrin Michel!
 Katrin Michel is a Postdoctoral Researcher at MIT and is fascinated by the question of how genes and proteins coordinate billions of neurons to form the functional networks of the human brain.
Katrin Michel is a Postdoctoral Researcher at MIT and is fascinated by the question of how genes and proteins coordinate billions of neurons to form the functional networks of the human brain.
Acknowledgement:
Through a partnership with genOway, we are able to distribute materials containing FLEx technology. Read the genOway press release for more information.
References
1. Atasoy, D., Y. Aponte, H. H. Su, and S. M. Sternson. 2008. “A FLEX Switch Targets Channelrhodopsin-2 to Multiple Cell Types for Imaging and Long-Range Circuit Mapping.” Journal of Neuroscience 28(28): 7025–30. PubMed PMID: 18614669. PubMed Central PMCID: PMC2593125.
2. Buchholz, F, P O Angrand, and A. Francis Stewart. 1998. “Improved Properties of FLP Recombinase Evolved by Cycling Mutagenesis.” Nature biotechnology 16(7): 657–62. PubMed PMID: 9661200.
3. Cardin, Jessica A. et al. 2009. “Driving Fast-Spiking Cells Induces Gamma Rhythm and Controls Sensory Responses.” Nature 459(7247): 663–67. PubMed PMID: 19396156. PubMed Central PMCID: PMC3655711.
4. Gronostajski, R M, and P D Sadowski. 1985. “The FLP Recombinase of the Saccharomyces Cerevisiae 2 Micron Plasmid Attaches Covalently to DNA via a Phosphotyrosyl Linkage.” Molecular and Cellular Biology 5(11): 3274–79. PubMed PMID: 2427927. PubMed Central PMCID: PMC369144.
5. Kranz, Andrea et al. 2010. “An Improved FLP Deleter Mouse in C57Bl/6 Based on FLPo Recombinase.” Genesis 48(8): 512–20. PubMed PMID: 20506501.
6. Logie, Colin, and A Francis Stewart. 1995. “Ligand-Regulated Site-Specific Recombination.” Proc. Natl. Acad. Sci. USA 92(June): 5940–44. PubMed PMID: 7597057. PubMed Central PMCID: PMC41617.
7. Metzger, D, J Clifford, H Chiba, and P Chambon. 1995. “Conditional Site-Specific Recombination in Mammalian Cells Using a Ligand-Dependent Chimeric Cre Recombinase.” Proc Natl Acad Sci U S A 92(15): 6991–95. PubMed PMID: 7624356. PubMed Central PMCID: PMC41457.
8. Schnütgen, Frank et al. 2003. “A Directional Strategy for Monitoring Cre-Mediated Recombination at the Cellular Level in the Mouse.” Nature Biotechnology 21(5): 562–65. PubMed PMID: 12665802.
9. Villa, Katherine L. et al. 2016. “Inhibitory Synapses Are Repeatedly Assembled and Removed at Persistent Sites In Vivo.” Neuron 89(4): 756–69. PubMed PMID: 26853302. PubMed Central PMCID: PMC4760889.
Additional Resources on the Addgene Blog
- Learn More about FLEX-vectors
- Learn More about Cre-Lox
- Check out Retrograde AAVs for Neurscience Research
Resources on Addgene.org
- Find Cre-Lox Plasmids
- Find Optogenetics Plasmids
- Find Chemogenetics Plasmids
Topics: Cre-lox, Plasmids, Neuroscience







Leave a Comment