Promoters control the binding of RNA polymerase and transcription factors. Since the promoter region drives transcription of a target gene, it therefore determines the timing of gene expression and largely defines the amount of recombinant protein that will be produced. Many commonly-used promoters, such as T7, CMV, EF1A, and SV40, are always active and thus referred to as constitutive promoters. Others are only active under specific circumstances. In a previous post, we discussed inducible promoters, which can be switched from a default OFF to an ON state, and how you might use these in your research. Today, we’ll look at repressible promoters, which can be switched from a default ON to an OFF state, as well as repressible binary systems. In recent years, a number of more complex systems have also been developed which don’t fit in either category, but those are a subject for a future post!
How are repressible promoters regulated?
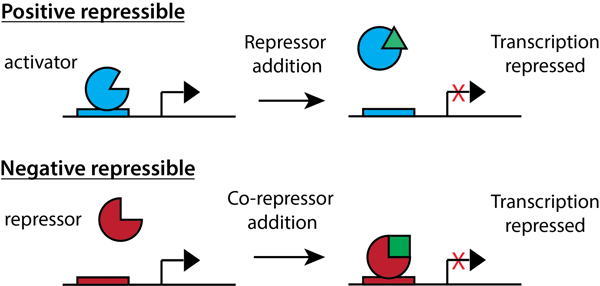
Like inducible promoters, repressible promoters can be regulated via positive or negative control.
Positive Repressible: An activator protein is bound, turning transcription ON. When a repressor binds the activator protein, the activator protein cannot bind the promoter sequence anymore and transcription is turned OFF.
Negative Repressible: A repressor protein is present, but cannot bind the promoter sequence and transcription is ON. Once a co-repressor protein binds the repressor protein, the repressor protein can bind to the operator. The bound repressor then prevents transcription from occurring, which means that transcription is now OFF.
We’ll look at examples of both natural and engineered repressible promoters.
Chemically repressible promoters
Tet-Off
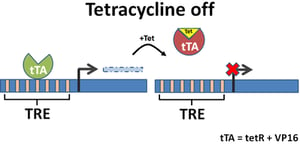
In its native context, the tetracycline repressor (TetR) can bind to the tetracycline operator sequences (TetO), preventing transcription. In the presence of tetracycline (Tet), TetR preferentially binds Tet over the TetO elements, allowing transcription to proceed (pJKR-H-tetR from the Church lab uses this system to control expression of GFP.)
To turn this inducible system into a repressible system, Gossen and Bujard created the tetracycline-controlled transactivator (tTA). They fused TetR with the transcriptional activation domain VP16 taken from the herpes simplex virus. tTA binds to promoters containing TetO elements (often linked in groups of seven as a Tet Response Element (TRE)), allowing transcription to proceed. When tetracycline or one of its derivatives is added, it binds tTA, resulting in a confirmation change that prevents binding to the promoter and turning transcription OFF.
Despite their bacterial origins, Tet systems function well in mammalian cells, and TRE-containing promoters can be used in the repressible manner described above, as well as the inducible manner detailed in our previous post.
In the past 30 years since the original Tet-Off system was described by Gossen and Bujard, both the promoter and transactivator components of the system have been engineered and improved. Takara’s “2nd generation Tet-Off vectors (Tet-Advanced)” are the most recent variants of the Tet-Off system. For more information on Addgene Tet plasmids, see our Tetracycline resource page.
Cumate Switch
Massie et. al developed the cumate-inducible gene expression system, which functions in a similar manner to the Tet system. The transactivator configuration of the system is a positive repressible promoter, and was engineered from the cmt and cym bacterial operons.
A chimeric transactivator, cTA, was created by fusing CymR to the activation domain VP16. cTA binds to promoters containing putative operator sequences (CuO) (linked in groups of 6), allowing transcription to proceed. When cumate is added, it binds cTA, resulting in a confirmation change that prevents binding to the promoter and turning transcription OFF.
The cumate switch system functions well in mammalian cells, and can be used in the repressible manner described above, as well as the inducible manner detailed in our previous post. While originally developed for use in mammalian cells, this system has also been implemented in a variety of other organisms, such as sphingomonads and bacillus.
ADH1
The ADH1 negative repressible promoter is commonly used in yeast. In the first 24 hours of culture, when glucose is abundant and ethanol concentrations are low, pADH1 displays low promoter activity (~20% activity of the strong yeast promoter pTEF). As ethanol accumulates, it binds to the repressor, enabling it to bind the promoter and repress pADH1 activity. Researchers have engineered pADH1 variants that are less sensitive to ethanol, including the “middle” ADH1 promoter pADH1m, which maintains activity during the late ethanol consumption phase of yeast culture.
Repressible Binary Systems
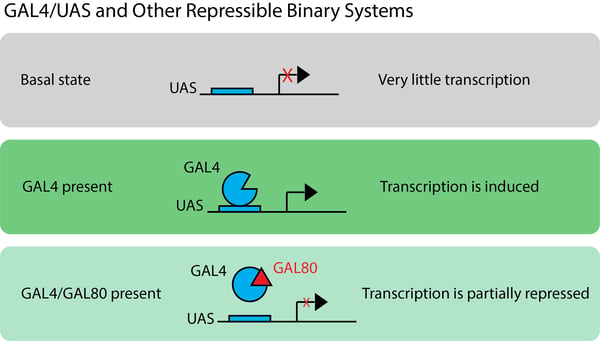
GAL4/UAS
In Drosophila or development studies, you may hear some transcription factor and promoter pairs referred to as binary systems. Binary systems are genetic tools that consist of two parts, both of which must be present for a gene of interest to be expressed. These permit exquisite control of gene expression and tracing of gene expression across development. One common binary system is the GAL4/UAS system isolated from yeast. In this system, UAS basal promoter expression is low but is activated by GAL4 binding to UAS.
If you place the GAL4 gene downstream of a tissue- or developmental stage-specific promoter and design a UAS reporter construct, the reporter will only be expressed where and/or when that promoter is active. In this way, you can interrogate the activity of uncharacterized promoters. Similarly, placing UAS upstream of a transgene permits directed expression of that gene in cells that also express GAL4. Usage of this system revolutionized genetic studies in flies.
An additional component, the GAL80 repressor, adds a repressible element to this binary system (making this a repressible binary system). The GAL80 repressor can bind to GAL4, partially inhibiting the binding of GAL4 to UAS. One can therefore place GAL4 and GAL80 under different promoters and create sophisticated patterns of UAS-driven gene expression. To further increase the complexity of the system, researchers have used split GAL4 architecture or combined the system with Flp/FRT, as reviewed by del Valle Rodriguez et al.
LexA/lexAop
A second binary system is LexA/lexAop, developed by Lai and Lee. LexA/lexAop is a complementary system to GAL4/UAS that functions in essentially the same manner, with LexA binding to the lexA operator (lexAop). Fusing the LexA DNA-binding domain (DBD) to GAL4 or VP16 activation domain results in chimeric GAL80-repressible and GAL80-insensitive activators of lexAop. This system is commonly used with GAL4/UAS to examine the expression of reporter genes, or to combine reporter expression from one promoter with transgene or siRNA expression from the other.
The Q system
Potter et al. from Liqun Luo’s lab developed the Q system from a gene cluster in the fungus Neurospora crassa. The QUAS promoter is less leaky than UAS in the basal state, and co-expression of the inducer QF can increase QUAS driven expression by ~3,300-fold in Drosophila cells and ~24,000-fold in human HeLa cells as compared to basal expression. Co-expression of repressor QS along with QF leads to intermediate expression from QUAS, as seen with GAL4/GAL80 and UAS. QF-mediated repression is reversible by the addition of quinic acid to Drosophila cells. Work by Wei et al. from Kang Shen’s lab has also shown that the Q system is also functional in C. elegans.
Subsequent work by the Luo and Potter labs has refined the Q system and introduced the second generation QF2 activators. These activators solve the problem of broad QF expression-induced lethality in flies. Riabinina et al. also developed chimeric transactivators GALQF and LexAQF, which activate their respective UAS and lexAop sequences but are also regulated by QS and quinic acid.
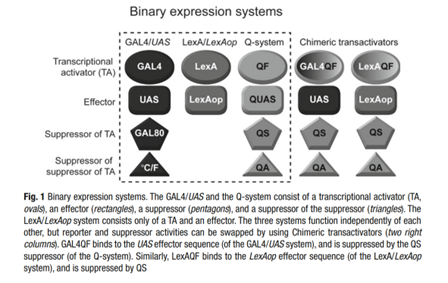 From Riabinia et. al.
From Riabinia et. al.
Using Binary Systems Together
You may be wondering, “why is it necessary to have all 3 of the different binary systems above if they all behave very similarly?” The simple answer is that sometimes it is necessary or desirable to control or look at multiple genes at the same time.
The 3 binary systems listed above are orthogonal, meaning that they do not affect each other. For example, the GAL4 transcription factor cannot drive transcription from the QUAS promoter, and the QF transcription factor cannot drive transcription from the UAS promoter. This is important because it allows them to be used together for complex genetic analysis in Drosophila or for precise control over multiple genes in synthetic systems. Recent work has even used all 3 in a single modular vector for ease of use!
Conclusion
This post contains only a brief overview of some of the most common repressible promoter systems, but there are certainly more out there! The repressible promoter systems detailed in this post can also be combined with other systems, such as inducible promoter systems, chemogenetic systems, and optogenetic systems, allowing for more complex control over desired gene expression. Take a look at all of the options and decide what might work best for your experiments!
This post was originally written by Mary Gearing in 2018, and updated by Hannah Dotson in Dec. 2022.
References and Resources
References
Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5547-51. doi: 10.1073/pnas.89.12.5547. PMID: 1319065; PMCID: PMC49329.
Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988 Jun;2(6):718-29. doi: 10.1101/gad.2.6.718. PMID: 2843425.
Takara Bio. Tet-On and Tet-Off systems: Second generation. https://www.takarabio.com/products/gene-function/tet-inducible-expression-systems/tet-systems-legacy-products/tet-on-and-tet-off-2nd-generation
Mullick, A., Xu, Y., Warren, R. et al. The cumate gene-switch: a system for regulated expression in mammalian cells. BMC Biotechnol 6, 43 (2006). https://doi.org/10.1186/1472-6750-6-43
Kaczmarczyk A, Vorholt JA, Francez-Charlot A. Cumate-inducible gene expression system for sphingomonads and other Alphaproteobacteria. Appl Environ Microbiol. 2013 Nov;79(21):6795-802. doi: 10.1128/AEM.02296-13. Epub 2013 Aug 30. PMID: 23995928; PMCID: PMC3811519.
Seo SO, Schmidt-Dannert C. Development of a synthetic cumate-inducible gene expression system for Bacillus. Appl Microbiol Biotechnol. 2019 Jan;103(1):303-313. doi: 10.1007/s00253-018-9485-4. Epub 2018 Nov 3. PMID: 30392122.
Pfeiffer, Barret D., et al. “Refinement of Tools for Targeted Gene Expression in Drosophila.” Genetics 186(2) (2010): 735-55. PubMed PMID: 20697123. PubMed Central PMCID: PMC2942869.
- Find plasmids from this publication at Addgene.
del Valle Rodriguez, Alberto, Dominic Didiano & Claude Desplan. “Power tools for gene expression and clonal analysis in Drosophila.” Nat Methods 9(1) (2011): 47-55. PubMed PMID: 22205518. PubMed Central PMCID: PMC3574576.
Lai, Sen-Li & Tzumin Lee. “Genetic mosaic with dual binary transcription systems in Drosophila.” Nat Neurosci 9(5) (2006): 703-9. PubMed PMID: 16582903.
Potter, Christopher J., et al. “The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis.” Cell 141(3) (2010): 536-48. PubMed PMID: 20434990. PubMed Central PMCID: PMC2883883.
- Find plasmids from this publication at Addgene.
Wei, Xing, Christopher J. Potter, Liqin Luo & Kang Shen. “Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans.” Nat Methods 9(4) (2012): 391-5. PubMed PMID: 22406855. PubMed Central PMCID: PMC3846601.
- Find plasmids from this publication at Addgene.
Riabinina, et al. “Improved and expanded Q-system reagents for genetic manipulations.” Nat Methods 12(3) (2015): 219-222. PubMed PMID: 25581800. PubMed Central PMCID: PMC4344399.
- Find plasmids from this publication at Addgene.
Riabinina O, Potter CJ. The Q-System: A Versatile Expression System for Drosophila. Methods Mol Biol. 2016;1478:53-78. doi: 10.1007/978-1-4939-6371-3_3. PMID: 27730575; PMCID: PMC5270762.
Wendler, F., Park, S., Hill, C. et al. A LexAop > UAS > QUAS trimeric plasmid to generate inducible and interconvertible Drosophila overexpression transgenes. Sci Rep 12, 3835 (2022). https://doi.org/10.1038/s41598-022-07852-7
De Boer, Herman A., Lisa K. Comstock & Mark Vasser. “The tac promoter: a functional hybrid derived from the trp and lac promoters.” Proc Natl Acad Sci U S A. 80(1) (1983): 21-5. PubMed PMID: 6337371. PubMed Central PMCID: PMC393301.
Da Silva, Nancy A. & Sneha Srikrishnan. “Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae.” FEMS Yeast Res. 12(2) (2012): 197-214. PubMed PMID: 22129153.
Additional Resources on the Addgene Blog
Resources at Addgene.org
Topics: Plasmid Elements, Plasmids 101, Plasmids






Leave a Comment