 This post was contributed by guest blogger Jon Chow, an immunology PhD student at Harvard University.
This post was contributed by guest blogger Jon Chow, an immunology PhD student at Harvard University.
In this second post in our quick guide to working with Drosophila, you’ll learn how to maniupate expression of your favorite gene (YFG) in flies. Read the first post here.
Once you’ve identified some fly stocks and other reagents of interest, the next question to ask is what to do with them. In some cases, there might be a mutation that disrupts the function of YFG. You could compare this mutant fly to one lacking the mutation in the same genetic background. In other cases, YFG or one of its mutant variants will need to be overexpressed or knocked down. To do this, Drosophila geneticists use the Gal4/UAS system. This incredibly useful, yet simple system allows you to systematically study gene function with temporal control and cell-type specificity!
The Gal4/UAS system
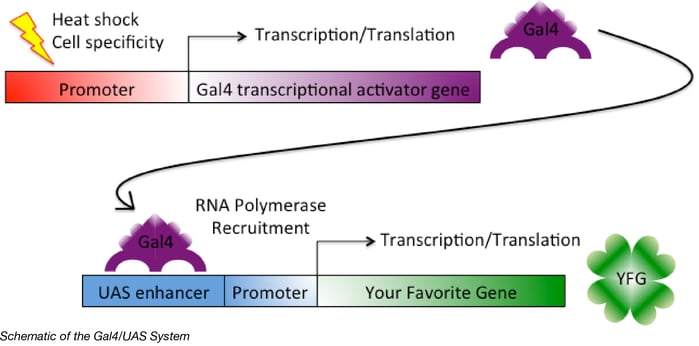
At a more detailed level, the Gal4/UAS system is a transcription activation system co-opted from yeast (1). It is a Drosophila geneticist’s main workhorse to turn genes on or off. Gal4 is a transcriptional activator that binds to UAS enhancer sequences found in DNA. It then recruits transcription machinery to the site to induce gene expression. Thus, genes and siRNA encoded downstream of the UAS sequence are only expressed when Gal4 is expressed.
Gal4 expression can be regulated by any chosen promoter, and Bloomington’s Stock Center has fly lines expressing Gal4 under a variety of promoters resulting in tissue specific expression. To express YFG in a specific tissue type, you mate your fly line containing YFG under the control of the UAS with a fly line expressing Gal4 in your tissue type of interest. You can quickly mate your UAS-YFG line with a variety of Gal4 lines and study gene expression in many different tissue types!
Although putting a gene of interest directly downstream of a tissue-specific promoter sounds simpler, this modular system saves you from having to create a new fly line every time you want to express YFG in a new tissue type (more details on making a fly line in the next blog post). For example, the hedgehog gene can be overexpressed in several cell types by using different Gal4 drivers. It is much easier to mate flies to get the right cell type expression as opposed to making a new fly with hedgehog under a new promoter each time a new hypothesis is tested.
Controlling multiple genes with Gal4/UAS system
One can also use Gal4 to drive expression of multiple genes under the control of UAS enhancer sequences. For example, if you discovered five new genes and wanted to express them within a specific neuronal compartment, you could express them individually or simultaneously. Do note for control purposes that there are dosage effects. For example, overexpression of two genes by one Gal4 driver will produce roughly half as much protein as overexpression of each gene individually.
Applications of the Gal4/UAS system
There are a number of fancy ways to use the Gal4/UAS system, and you can read more about them in review articles (2–4). You can also take a look at my recent paper to see the Gal4/UAS system in action (5). I use the Gal4/UAS system to knock down gene expression, rescue knock out mutations, and overexpress viral genes. Additionally, other modular expression systems exist (eg. LexA/LexAop and Cre/Lox systems) in case you want to express one gene in one cell type and another gene in a different cell type (6–8).
Although it’s often very easy to get GAL4 UAS experiments up and running with fly lines that are already available, sometimes you’ll have to make your own. In my next post, I’ll explain the relatively straightforward process of generating your own fly lines. In the meantime, be sure to browse the publication references and web links from my first post and start designing your first fly experiment!
Many thanks to our guest blogger Jon Chow.
 Jon Chow is finishing his immunology PhD at Harvard University. He has had nearly a decade of experience working with Drosophila as an experimental system. Jon enjoys studying the never-ending struggle between pathogens and their hosts. It’s not always clear which side he roots for. You can try to convince him to use Twitter @jonchowphd but he still reads “#” as a “pound symbol”.
Jon Chow is finishing his immunology PhD at Harvard University. He has had nearly a decade of experience working with Drosophila as an experimental system. Jon enjoys studying the never-ending struggle between pathogens and their hosts. It’s not always clear which side he roots for. You can try to convince him to use Twitter @jonchowphd but he still reads “#” as a “pound symbol”.
References
1. A. H. Brand, N. Perrimon, Targeted gene expression as a means of altering cell fates and generating dominant phenotypes., Development 118, 401–15 (1993). PubMed PMID: 8223268.
2. J. B. Duffy, GAL4 System in Drosophila : A Fly Geneticist’s Swiss Army Knife, Genesis 34, 1–15 (2002).PubMed PMID: 12324939.
3. S. E. McGuire, G. Roman, R. L. Davis, Gene expression systems in Drosophila: A synthesis of time and space, Trends Genet. 20, 384–391 (2004). PubMed PMID: 15262411.
4. T. D. Southall, D. A. Elliott, A. H. Brand, The GAL4 System : A Versatile Toolkit for Gene Expression in Drosophila, 3, 1–10 (2015). PubMed PMID: 21356876.
5. J. Chow, Z. Marka, I. Bartos, S. Marka, J. C. Kagan, Environmental Stress Causes Lethal Neuro-Trauma during Asymptomatic Viral Infections, Cell Host Microbe 22, 48-60 (2017). PubMed PMID: 28704652.
6. V. Rodríguez, D. Didiano, C. Desplan, Power tools for gene expression and clonal analysis in Drosophila, 9, 47–55 (2012). PubMed PMID: 22205518. PubMed Central PMCID: PMC3574576.
7. S.-L. Lai, T. Lee, Genetic mosaic with dual binary transcriptional systems in Drosophila., Nat. Neurosci. 9, 703–709 (2006). PubMed PMID: 16582903.
8. C. J. Potter, B. Tasic, E. V Russler, L. Liang, L. Luo, The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis., Cell 141, 536–48 (2010). PubMed PMID: 20434990. PubMed Central PMCID: PMC2883883.
Topics: Drosophila, Other






Leave a Comment