Plasmids designed to express genes in a given host cell type are generally broken down into two broad categories, prokaryotic or eukaryotic, based on the functional elements they contain. Plasmid DNA in both prokaryotic and eukaryotic systems must be transcribed into RNA, which occurs in three phases: initiation, elongation, and termination. In a previous post we discussed the promoter's role in the initiation step of gene transcription; today we'll provide an overview on how transcription stops, or termination. Read on to learn more!
Download our Plasmids 101 eBook to get all the background you need to start working with plasmids!
What are termination and polyadenylation?
The role of the terminator, a sequence-based element, is to define the end of a transcriptional unit (such as a gene) and initiate the process of releasing the newly synthesized RNA from the transcription machinery. Terminators are found downstream of the gene to be transcribed, and typically occur directly after any 3’ regulatory elements, such as the polyadenylation or poly(A) signal. While many studies focus on promoter strength as a determinant of gene expression levels, the terminator also plays an important role in RNA processing and contributes to variability in RNA half-life, and ultimately gene-expression.
Polyadenylation is the post-transcriptional additional of multiple adenine (A) nucleotides to the tail of a messenger RNA transcript. The purpose and mechanism of polyadenylation vary across cell types, but polyadenylation generally serves to promote transcript longevity in eukaryotes and promote transcript degradation in prokaryotes.
Prokaryotic termination
Prokaryotic termination mechanisms fall under two general categories: rho-dependent and rho-independent. Rho factor is a helicase which assists RNA polymerase in the termination of the transcript. Rho-dependent terminators are not usually employed in plasmid-based expression systems, so these will not be detailed here, but additional references are provided at the end.
Nearly all common bacterial expression plasmids use Rho-independent terminators, which include naturally occurring terminators, such as T7 and rrnB, as well as engineered high-efficiency terminators such as T0. Rho-independent termination is also known as intrinsic termination, and it relies on the formation of a GC-rich hairpin in the RNA transcript followed by a weakly bound poly-uracil tract as shown in the figure to the right. The tertiary structure of the hairpin-DNA complex is thought to destabilize the transcription complex, initiating cleavage of the transcript.
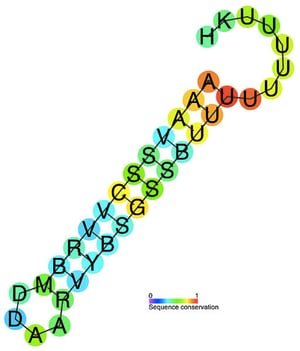 |
| Figure 1: A predicted conserved secondary structure and sequence conserved Rho-independent termination annotation for 90 bacterial elements. By Ppgardne at English Wikipedia, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=39322032 |
No terminator is 100% efficient at halting transcription of the template and initiating the desired cleavage event, although some engineered terminators come close (>95%). For most purposes, however, any common terminator will suffice. Many commercial expression vectors use double terminators to reduce unwanted translation of downstream elements. A high affinity terminator may be desired for multi-cistronic constructs where high termination efficiency is necessary to minimize transcriptional read-through. Chris Voigt’s lab has characterized a set of prokaryotic terminators and deposited several with Addgene (1).
Prokaryotic polyadenylation
Although mostly thought of as a eukaryotic-specific process, prokaryotes also add poly(A) tails to certain RNAs. Unlike the eukaryotic mechanism which requires a consensus sequence for the addition of a poly(A) tail, the addition of a poly(A) tail on a prokaryotic transcript is non-specific and can be added to any accessible 3' end. The presence of the poly(A) tail targets the RNA to the degradosome, which contains enzymes that cut RNA not protected by secondary structure. It is thought that poly(A)s are used to control the cellular concentration of regulatory RNAs and may additionally act as a quality control mechanism to rid the cell of mis-folded RNAs.
Eukaryotic termination and polyadenylation
Unlike prokaryotes that have a single RNA polymerase for transcription, eukaryotes have three RNA polymerases (Polymerases I, II, and III), each responsible for transcribing different types of RNA: Polymerase I is responsible for ribosomal RNA, Polymerase II is responsible for mRNA and miRNAs, and Polymerase III transcribes tRNA and other short RNAs. Although not as well studied as prokaryotic termination, the basic processes for eukaryotic termination are understood and it has been noted that each eukaryotic RNA polymerase terminates differently. Polymerase III, for example, relies on a specific sequence and RNA secondary structure to induce transcript cleavage, similar to the Rho-independent termination found in prokaryotes. This is different than Polymerases I and II, which both require binding of termination factors. Although both are termination factor dependent, Polymerases I and II employ different mechanisms to terminate transcription. Polymerase I uses a process similar to the prokaryotic Rho-dependent mechanism, whereas Polymerase II termination is more complex and involves two RNA polymerase-associated proteins, CPSF and CstF, which are responsible for recruiting the cleavage and polyadenylation enzymes, in a process that seems to couple termination with polyadenlyation.
Mammalian expression plasmids are primarily used to create mRNA and the commonly used mammalian terminators (SV40, hGH, BGH, and rbGlob) include the sequence motif AAUAAA which promotes both polyadenylation and termination. Out of those listed, the SV40 late polyA and rbGlob polyA are thought to be more efficient in terminating transcription due to the presence of additional helper sequences (2-3).
As alluded to above, termination and polyadenylation of Polymerase II transcripts (and therefore mRNAs) are coordinated processes. Cleavage between the consensus motif and a downstream GU-rich region (shown in the figure below) releases the mRNA from the polymerase and creates a free 3' end which is now available for polyadenylation. The addition of the poly(A) tail is important for stability of the mRNA, protection from degradation, and is integral to the nuclear export and translation processes as well.
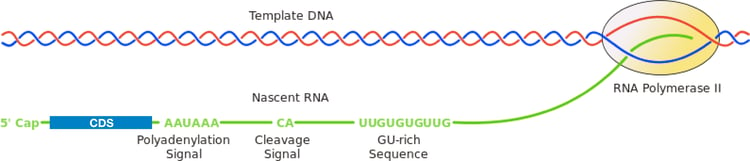 |
| Figure 2: The conserved eukaryotic polyadenylation signal directs cleavage at the cleave signal and addition of a poly-A tail to the mRNA transcript. By Arunreginald at en.wikipedia, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=17554393 |
Use of mammalian poly(A) signals in combination with certain viral packaging systems has been associated with reduced viral titer, but improved transcript life, and so should be handled with care (4). For this reason, viral vectors often employ other non-poly(A) transcript-stabilizing and nuclear export elements such as WPRE (5) and CTE (6), and/or use weaker poly(A) signals like BGH.
References
- Peters JM, Vangeloff AD, Landick R. Bacterial Transcription Terminators: The RNA 3′-End Chronicles. Journal of molecular biology. 2011. PMID: 21439297. PubMed Central PMCID: PMC3622210.
- Schek N, Cooke C, Alwine JC. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Molecular and Cellular Biology. 1992. PMID: 1333042. PubMed Central PMCID: PMC360476.
- Gil A, Proudfoot NJ. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3' end formation. Cell. 1987. PMID: 3568131.
- Hager S, Frame FM, Collins AT, Burns JE, Maitland NJ. An Internal Polyadenylation Signal Substantially Increases Expression Levels of Lentivirus-Delivered Transgenes but Has the Potential to Reduce Viral Titer in a Promoter-Dependent Manner. Hum Gene Ther. 2008. PMID: 18627247
- Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element Enhances Expression of Transgenes Delivered by Retroviral Vectors. Journal of Virology. 1999. PubMed PMID: 10074136. PubMed Central PMCID: PMC104046.
- Wodrich H, Schambach A, Kräusslich H-G. Multiple copies of the Mason–Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Research. 2000. PubMed PMID:10648781. PubMed Central PMCID: PMC102582.
Additional Resources
- A detailed analysis of yeast terminators of varying strength.
- A discussion of the effects of various terminators in the context of AAV transgene expression levels.
- Transcription Termination
- Rho-dependent termination
- DNA transcription
Polyadenylation:
Resources on the Addgene Blog
- Learn how the Promoter Controls Gene Expression
- Learn about New Cloning Techniques
- Read all of our Plasmids 101 Posts
Resources on Addgene.org
- Browse our Plasmid Reference Pages
- Just Starting Work in a New Field? Check out Our Plasmid Collections!
- Join the Addgene Community
Topics: Plasmid Elements, Plasmids 101, Plasmids







Leave a Comment