Understanding the characteristics of natural plasmids and how they function in nature can inform us on how to create better recombinant vectors. In this blog post, we’ll define plasmid host range, identify a few of the features that confer broad host range in plasmids in nature, and provide tips on how we can harness those features to create broad host range recombinant vectors.
A plasmid’s host range is determined by the different hosts that a plasmid can transfer to, replicate in, and persist in. Some plasmids have a narrow host range, while others have a broad host range and can replicate and stably maintain the genes they carry among organisms belonging to many different phylogenetic groups (Jain and Srivastava 2013).
Recombinant vectors that are made from broad host range plasmids are more stable, can be easily cloned in different bacteria, and can be transferred to various experimental hosts (Jain and Srivastava 2013). They are also easily-shareable tools within the scientific community, as they allow for simplified transformation processes, and can be used by scientists across disciplines.
What confers broad host range in plasmids?
Not all plasmids were created equally. Only a handful of plasmids have been identified as broad host range plasmids. The properties that influence host range are not fully understood, but scientists have identified some of the features that confer broad host range. These features are:
- The plasmid origin of replication (ori) contains structural elements that are versatile and adaptable. Variation in ori structure allows plasmids to utilize diverse host replication machinery. For instance, the ori typically contains iterons that serve as recognition sites for replication initiation proteins. These iterons vary in their sequence and number and can alter the efficiency of replication in different bacterial hosts (Doran et al. 1998). Regulatory elements within the ori also control copy number and stability, which influence the plasmid’s ability to establish and maintain itself within host cells.
- The plasmid has multiple ori that are functionally differentiated from one another. Based on their structural elements, oris interact with various RNA polymerases and proteins from bacterial hosts. In a plasmid with multiple oris, one may function in one type of host and the others are functional in other hosts. For example, the pJD4 plasmid harbors distinct origins (ori1, ori2, ori3) and replication initiation proteins (RepA, RepB) necessary for replication in various hosts (Pagatto and Dillon 2001).
- The plasmid has its own replication initiation factors and proteins independent of the host. Host range depends on whether the host has the components to recognize the ori and allow for replication. However, if plasmids have their own replication machinery such as Rep or other initiator proteins, they do not require host proteins for replication and can maintain themselves in many different bacteria (Meyer 2009; Jain and Srivastava 2013).
- The plasmid does not have many restriction enzyme sites. Hosts often have a restriction barrier as a defense mechanism, and they can degrade foreign bacteria through the use of their restriction enzymes (Meyer et al. 1977). So if a plasmid has few restriction sites, it may be able to sneak past this checkpoint.
The plasmid is self-transmissible, meaning that the plasmid is conjugative and carries the genes necessary for transfer initiation between bacterial hosts. Also, self-transmissible plasmids may have high copy numbers and reduced metabolic load (Meyer 2009).
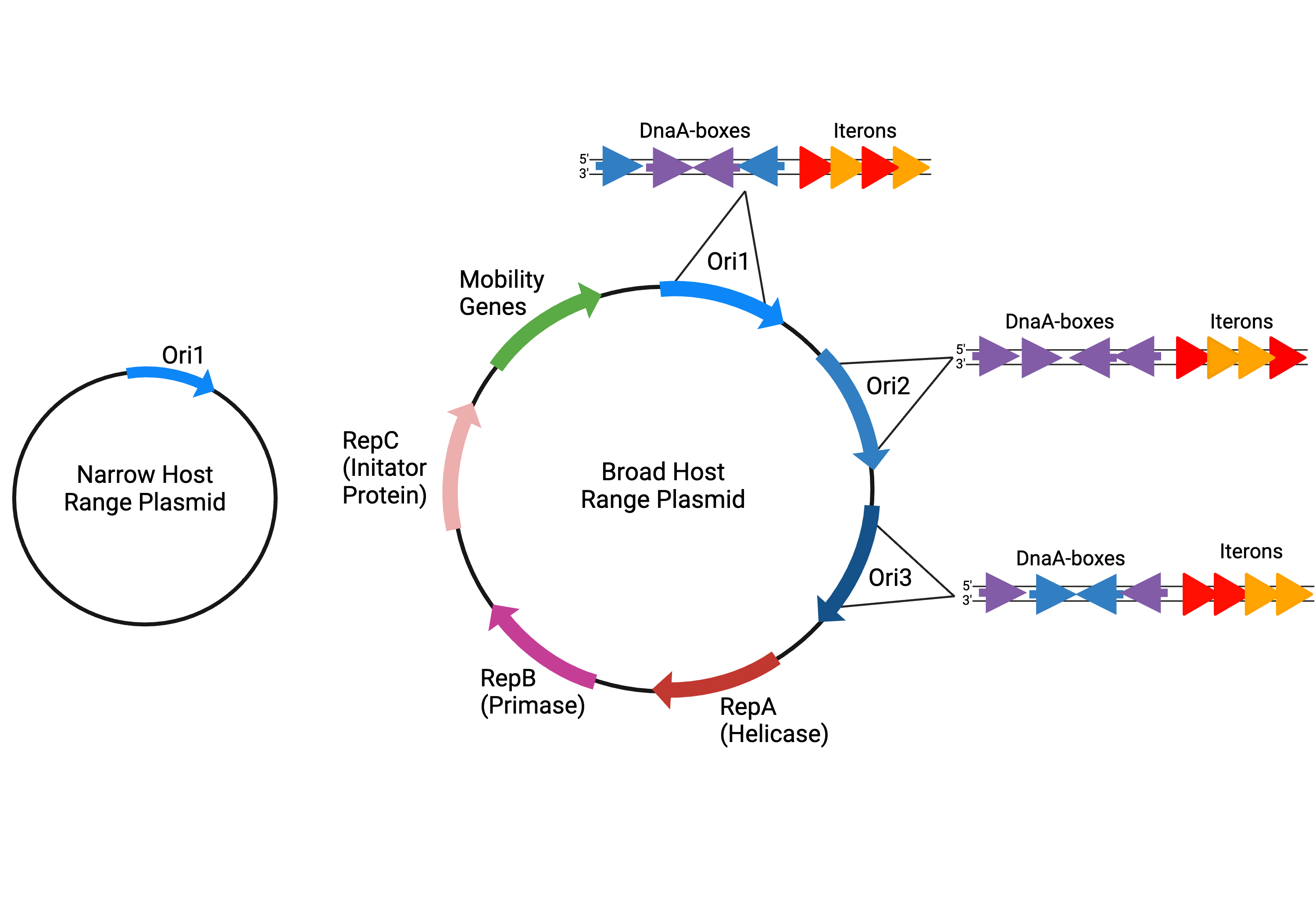 |
| Figure 1: Idealized examples of a narrow host range plasmid with only one ori and a broad host range plasmid with multiple oris that vary in DnaA-box and iteron structure, host-independent |
Building broad host range recombinant vectors
Replicons, which consist of a plasmid’s ori and all of its control elements, from broad host range plasmids can be used to create recombinant vectors that can replicate in different bacteria and express proteins within different host organisms. pBBR1-based plasmids and plasmids in incompatibility groups N, P, Q, and W are known to have replicons with broad host ranges. Popular replicons from these groups include: RK2 (IncP), RSF1010 (IncQ), and pSa (IncW) (Schmidhauser et al. 1988; Lale et al 2011; Jain and Srivastava 2013). Conveniently, these broad host range plasmids all replicate in E. coli, which is the most commonly used bacteria in lab manipulations. Table 1 has a detailed list of the bacterial species these plasmids are compatible with.
Table 1: Non-exhaustive list of common broad host range plasmids and the bacterial species in which they are known to replicate. Information sourced from Lale et al 2011 and Jain and Srivastava 2013.
|
Family |
Replicon |
Compatible with |
Shown to work in |
|
IncN |
pCU1 |
Gram-negative |
Agrobacterium spp., Bradyrhizobium spp., Rhizobium spp. |
|
IncP |
RK2 |
Gram-negative |
Achromobacter parvulus, Acinetobacter spp., Aeromonas spp., Agrobacterium spp., Alcaligenes spp., Aliivibrio salmonicida, Anabaena spp., Azospirillum braziliense, Azotobacter spp., Bordetella spp., Caulobacter spp., Enterobacteriaceae, Gluconacetobacter xylinus, Haemophilus influenzae, Hypomycrobium X, Legionella pneumophila, Methylophilus methyltrophus, Methylococcus methanolicus, Methylosinus trichosporium, Myxococcus xanthus, Neisseria spp., Paracoccous denitrificans, Pseudomonas spp., Rhizobium spp., Rhodopseudomonas spp., Rhodospirillum spp., Shewanella spp., Thiobacillus spp., Xanthomonas campestris |
|
IncQ |
RSF1010, R300B, R1162 |
Gram-negative and Gram-positive |
Acinetobacter calcoaceticus, Actinobacillus pleuropneumoniae, Actinomyces naeslundii, A. viscosus, Aerobacter aerogenes, Aeromonas hydrophila, Agrobacterium tumefaciens, Alcaligenes eutrophus, Azotobacter vinelandii, Brevibacterium methylicum, Caulobacter crescentus, Desulfovibrio vulgaris, Erwinia carotovora, E. chrysanthemi, Escherichia coli, Gluconacetobacter xylinus, Gluconobacter spp., Hyphomicrobium spp., Klebsiella aerogenes, K. pneumoniae, Methylophilus methylotrophus, Moraxella spp., Mycobacterium aurum, M. smegmatis, Paracoccus denitrificans, Pasteurella multocida, Porphyromonas gingivalis, Proteus mirabilis, Providencia spp., Pseudomonas spp., Rhizobium leguminosarum, R. meliloti, Rhodobacter sphaeroides, R. capsulatus, Rhodopseudomonas spheroides, Salmonella spp., Serratia marcescens, Streptomyces lividans, Synechococcus spp., Thiobacillus ferrooxidans, Vibrio salmonicida, Yersinia enterocolitica, Xanthomonas campestris, X. maltophilia |
|
IncW |
pSa, pR388 |
Gram-negative |
Acinetobacter calcoaceticus, Aeromonas liquefaciens, A. salmonicida, Agrobacterium tumefaciens, A. rhizogenes, Alcaligenes eutrophus, Enterobacter sp., Erwinia amylovora, E. carotovora subsp. Carotovora, E. herbicola, E. rubrifaciens, E. stewartii, Escherichia coli, Klebsiella spp., Legionella pneumophila, Methylophilus methylotrophus, Myxococcus virescens, M. xanthus, Proteus rettgeri, P. mirabilis, Providencia stuartii, Pseudomonas spp., Rhizobium leguminosarum, R. trifolii, Salmonella enteritidis, S. typhimurium, S. ordonez, Serratia marcescens, Shigella spp., Vibrio cholerae, Xanthomonas campestris pv. campestris, X. campestris pv. malvacearum, Zymomonas mobilis |
|
pBBR1 based |
pBBR1 |
Gram-negative |
Alcaligenes eutrophus, Bartonella bacilliformis, Bordetella spp., Brucella spp., Caulobacter crescentus, Escherichia coli, Gluconacetobacter xylinus, Paracoccous denitrificans, Pseudomonas fluorescens, P. putida, Rhizobium meliloti, R. leguminosarum by. viciae, Rhodobacter sphaeroides, Salmonella typhimurium, Vibrio cholerae, Xanthomonas campestris |
In addition to incorporating broad host range replicons, recombinant vectors can be modified further to increase the range of hosts in which proteins can be expressed. For example, scientists can incorporate interchangeable antibiotic resistance cassettes (Prior et al. 2010), inducible promoters with different degrees of stringency and inducibility (Toh et al. 2023), and additional cloning sites (Keen 1988).
Find broad host range recombinant vectors at Addgene!
References and resources
References
Doran, K. S., Konieczny, I., & Helinski, D. R. (1998). Replication Origin of the Broad Host Range Plasmid RK2: Positioning of Various Motifs is Critical for Initiation of Replication. Journal of Biological Chemistry, 273(14), 8447–8453. https://doi.org/10.1074/jbc.273.14.8447
Jain, A., & Srivastava, P. (2013). Broad host range plasmids. FEMS Microbiology Letters, 348(2), 87–96. https://doi.org/10.1111/1574-6968.12241
Keen, N. T., Tamaki, S., Kobayashi, D., & Trollinger, D. (1988). Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene, 70(1), 191–197. https://doi.org/10.1016/0378-1119(88)90117-5
Lale, R., Brautaset, T., & Valla, S. (2011). Broad-host-range plasmid vectors for gene expression in bacteria. Methods in Molecular Biology (Clifton, N.J.), 765, 327–343. https://doi.org/10.1007/978-1-61779-197-0_19
Meyer, R. (2009). Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid, 62(2), 57–70. https://doi.org/10.1016/j.plasmid.2009.05.001
Meyer, R., Figurski, D., & Helinski, D. R. (1977). Physical and genetic studies with restriction endonucleases on the broad host-range plasmid RK2. Molecular & General Genetics: MGG, 152(3), 129–135. https://doi.org/10.1007/BF00268809
Prior, J. E., Lynch, M. D., & Gill, R. T. (2010). Broad-host-range vectors for protein expression across gram negative hosts. Biotechnology and Bioengineering, 106(2), 326–332. https://doi.org/10.1002/bit.22695
Schmidhauser, T. J., Ditta, G., & Helinski, D. R. (1988). Chapter 15—Broad-Host-Range Plasmid Cloning Vectors for Gram-Negative Bacteria. In R. L. Rodriguez & D. T. Denhardt (Eds.), Vectors (pp. 287–332). Butterworth-Heinemann. https://doi.org/10.1016/B978-0-409-90042-2.50021-0
Toh, W. K., Teo, Y. L., Tor, X. Y., Loh, P. C., & Wong, H. L. (2023). Development of constitutive and IPTG-inducible integron promoter-based expression systems for Escherichia coli and Agrobacterium tumefaciens. 3 Biotech, 13(3), 91. https://doi.org/10.1007/s13205-023-03507-0
More resources on the Addgene blog
Plasmids 101: Origin of Replication
Plasmids 101: What is a Plasmid?
Plasmids 101: Transformation, Transduction, Bacterial Conjugation, and Tranfection
Topics: Plasmids 101, Plasmids






Leave a Comment