Plasmid incompatibility is defined as the inability of different plasmids to be maintained in one bacterial cell. In this Plasmids 101 post, we’ll cover why this happens, how it might affect your work, and how understanding it can be used for good.
First, why are plasmids incompatible? Plasmid incompatibility occurs when multiple plasmids within one cell have the same replicon and/or partitioning system. Let’s start with the replicon- the part of the plasmid that contains the origin of replication and the replication control machinery (Need a refresher on the Origin of Replication? See our Plasmids 101: Origin of Replication blog).
The replication problem
Plasmids with the same replicon are incompatible because they compete for the same replication control machinery within the cell. Most plasmids encode a negative regulation system involving antisense RNAs or iterons that inhibits replication when copy number in the cell is high, but allows replication when copy number in the cell falls too low (Novcik, 1987).
Antisense RNAs encoded on the plasmid either indirectly inhibit replication through inhibiting translation of replication machinery proteins, or directly inhibit replication by binding to the origin of replication and blocking replication machinery (del Solar, 1998). As the copy number of the plasmid increases, increased amounts of these antisense RNAs inhibit plasmid replication.
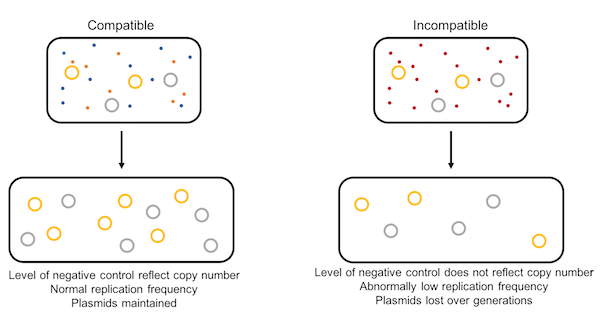 |
| Figure 1: The replication problem. |
Iterons are repeated sequences within the origin of replication and are required for replication in plasmids that contain them (del Solar et al., 1998). There are two models for iteron-based plasmid replication control that are not necessarily mutually exclusive. In the first, the replication initiation factor RepA binds to the iteron to initiation replication. However, in iteron-containing plasmids, RepA can also bind to its own promoter and inhibit its own transcription. As copy number increases, RepA transcription is repressed and replication is downregulated. In the second model, termed the “handcuff” model, iterons of two separate plasmids bind to the same RepA molecules, forming a “handcuff” structure that physically blocks replication machinery when plasmid copy number is high (del Solar et al., 1998).
In cases of compatible plasmids, different negative regulation systems control plasmid replication of each plasmid using unique replication machinery. However, when plasmids have the same origin of replication, the negative regulation system cannot distinguish between the different plasmids. The concentration of negative regulation system elements in the cell is artificially high for both plasmids, so each plasmid “thinks” it has a higher copy number than it actually does, and neither plasmid is maintained (Figure 1).
The partitioning problem
Plasmid incompatibility also occurs because of plasmid partitioning (Schumacher, 2012). This concept is a little more complex than simply competing for replication control factors and tends to be more of a concern with low copy plasmids, though the partitioning problem does occur in high copy plasmids (Diaz et al., 2015). Plasmids contain a system to partition themselves to each daughter cell during cell division. They don’t simply rely on chance (or random diffusion) to make this happen.
There are several different versions of the plasmid partitioning system in the bacteria, but generally it consists of a centromere-like region on the plasmid, a centromere-like region binding protein (the CBP), and a partition NTPase (Schumacher, 2012). During cell division, CBPs bind to the centromere-like region on each plasmid and pair the plasmids together (akin to sister chromatids coming together in eukaryotic cells). The partition NTPase is recruited and “walks” each plasmid to a separate daughter cell. When plasmids are compatible, different CBPs bind to each plasmid type, and different NTPases separate the plasmid pairs into the new daughter cells.
For high copy plasmids, incompatibility due to partitioning is similar to incompatibility due to having the same replication machinery. High copy plasmids with the same centromere-like binding region compete for the same CBPs and NTPase to correctly partition plasmids to each daughter cell. When there are not enough CBPs and NTPase to go around, the plasmids are randomly positioned, leading to plasmid loss (Diaz et al., 2015).
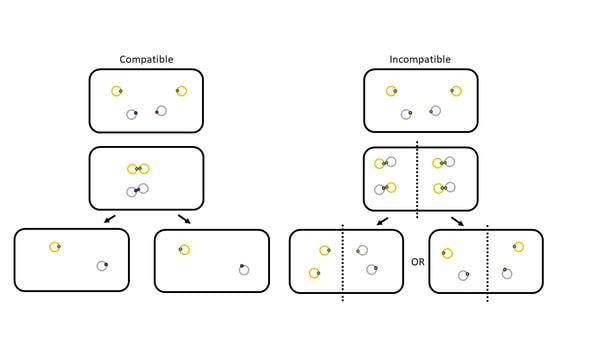 |
| Figure 2: The partitioning problem. |
For low copy plasmids, the main theory for plasmid incompatibility is due to an identification issue: the CBPs in the cell can’t tell the difference between plasmids if they have the same centromere-like region, and the NTPases end up “walking” the same plasmids to one daughter cell, instead of partitioning each type of plasmid into a separate daughter cells (Figure 2) (Ebersbach et al., 2005). However, more recent studies suggest that the late replication that occurs for some types low copy plasmids during cell division simply does not allow for enough time for correct partitioning to occur (Diaz et al., 2015).
It is also worth noting that plasmids can be symmetrically incompatible, meaning that both plasmids are lost from the cell with the same probability, or asymmetrical, where one plasmid lost from the cell at a higher rate than the coresident plasmid. Asymmetric plasmid loss occurs for several reasons, including one plasmid blocking replication of another and one plasmid outcompeting the other plasmid for replication or partitioning machinery (Novick, 1987). In cases where one plasmid outcompetes the other, usually a smaller, higher copy plasmid can more effectively titrate replication or partitioning machinery away from a larger, low copy plasmid and the larger, low copy plasmid is more likely to be lost over time.
Is plasmid incompatibility going to affect my cloning project?
If you want to clone two plasmids within the same bacterial cell, think about whether plasmid incompatibility will affect your plans. This situation may arise for a microbiologist, where cloning two plasmids into one bacterium is not uncommon, or perhaps a synthetic biologist who wants to develop a plasmid-based tool that is easy to use with other plasmids.
How do you know if your plasmids are incompatible? Scientists have developed a system of bacterial incompatibility groups based on similarity of replication and partitioning systems where the plasmids belonging to the same group are incompatible. Currently, there are 27 incompatibility (Inc) groups in the Enterobacteriaceae family alone, and this is expanding as we obtain more genomic data about plasmids (Shintani et al., 2015). Now, bioinformatics tools help researchers determine how compatible their plasmids will be based on sequence data. However, if these do not provide clarity, a wet lab transformation experiment will ultimately inform whether your plasmids are compatible or not.
Using plasmid incompatibility for therapeutic advantage
Although it may seem like plasmid incompatibility may be standing in your way to bacterial cloning glory, scientists have applied these concepts to deal with the more sinister plasmids of the microbiological world. (Want to know more about these plasmid types? Read our Plasmids 101: Environmental Plasmids blog). Scientists have designed small, high copy incompatible plasmids that lead to asymmetrical plasmid loss of the usually large, low copy virulence plasmids. This strategy has been successful in displacing, or “curing” the virulence plasmids from the bacterial pathogens Yersinia pestis, Agrobacterium tumefaciens, and Bacillus anthracis (Lui et al., 2012, Ni et al., 2008, Uraji et al., 2002). Scientists are beginning to use plasmid incompatibility to combat antimicrobial resistance. Using novel plasmids based partly on plasmid incompatibility with antibiotic-resistance plasmids, scientists have expelled antibiotic resistant plasmids from Enterobacteriaceae family members within the mouse gut (Kamruzzaman et al., 2017).
Additional resources on the Addgene blog
- Read all of our Plasmids 101 blog posts
- Learn more about the origin of replication
- Find blog posts about plasmid cloning
Resources on Addgene.org
- Download the Plasmids 101 eBook
- Browse plasmids at Addgene
- Read our Molecular Biology Reference
References
del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R (1998) Replication and Control of Circular Bacterial Plasmids. Microbiology and Molecular Biology Reviews 62(2): 434–464. Pubmed PMID: 9618448
Diaz R, Rech J, Bouet J-Y (2015) Imaging centromere-based incompatibilities: Insights into the mechanism of incompatibility mediated by low-copy number plasmids. Plasmid 80:54–62 . https://doi.org/10.1016/j.plasmid.2015.03.007 Pubmed PMID: 25889267
Ebersbach G, Sherratt DJ, Gerdes K (2005) Partition-associated incompatibility caused by random assortment of pure plasmid clusters. Molecular Microbiology 56:1430–1440 . https://doi.org/10.1111/j.1365-2958.2005.04643.x Pubmed PMID: 15916596
Kamruzzaman M, Shoma S, Thomas CM, Partridge SR, Iredell JR (2017) Plasmid interference for curing antibiotic resistance plasmids in vivo. PLOS ONE 12:e0172913 . https://doi.org/10.1371/journal.pone.0172913 Pubmed PMID: 28245276
Liu X, Wang D, Wang H, Feng E, Zhu L, Wang H (2012) Curing of Plasmid pXO1 from Bacillus anthracis Using Plasmid Incompatibility. PLoS ONE 7:e29875 . https://doi.org/10.1371/journal.pone.0029875 Pubmed PMID: 22253811
Ni B, Du Z, Guo Z, Zhang Y, Yang R (2008) Curing of four different plasmids in Yersinia pestis using plasmid incompatibility. Letters in Applied Microbiology 47:235–240 . https://doi.org/10.1111/j.1472-765x.2008.02426.x Pubmed PMID: 19241516
Novick RP (1987) Plasmid incompatibility. Microbiology Reviews. 1987;51(4):381–395. Pubmed PMID: 3325793
Schumacher MA (2012) Bacterial plasmid partition machinery: a minimalist approach to survival. Current Opinion in Structural Biology 22:72–79 . https://doi.org/10.1016/j.sbi.2011.11.001 Pubmed PMID: 22153351
Shintani M, Sanchez ZK, Kimbara K (2015) Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Frontiers in Microbiology 6: . https://doi.org/10.3389/fmicb.2015.00242 Pubmed PMID: 25873913
Uraji M, Suzuki K, Yoshida K (2002) A novel plasmid curing method using incompatibility of plant pathogenic Ti plasmids in Agrobacterium tumefaciens. Genes & Genetic Systems 77:1–9 . https://doi.org/10.1266/ggs.77.1 Pubmed PMID: 12036099
Topics: Plasmids 101, Plasmids






Leave a Comment