You’ve spent days and weeks thinking of an amazing project. You’ve written your protocols, designed your experiments, and prepared your reagents. You’re going to engineer the best thing since CRISPR; you are ready to clone! But...how?
Cloning is no longer limited to restriction enzymes - although many labs still love using them! With so many technologies currently available, you now face the challenge of trying to figure out what system might work best for your particular purposes. We’re here to help with that!
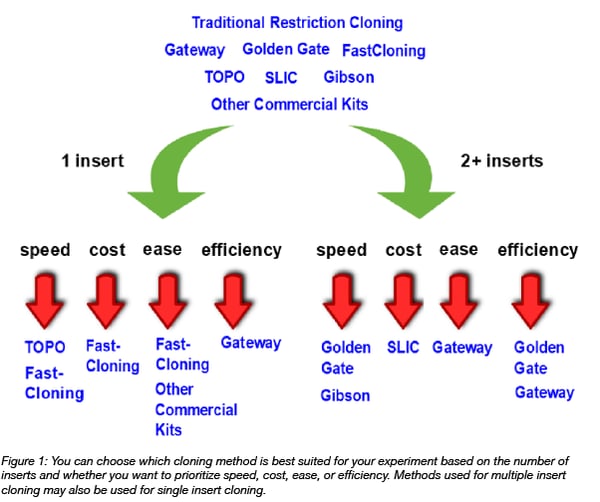
Four important factors to consider for any experiment are:
- Speed
- Cost
- Difficulty
- Efficiency
For cloning in particular, another important consideration is:
- The number of inserts
...as some methods are more amenable to combining multiple DNA fragments than others.
In today’s post, we will focus on a high-level comparison of popular cloning methodologies, with a special emphasis on these five categories. Please note that many of the following cloning methodologies have been extensively described in other articles; so if you are interested in learning more about any one of them, please click on the links throughout the text or see the “Additional Resources” section at the end of the post!
Speed:
Your grant deadline is approaching or your advisor is demanding results. If you’re in a hurry, Golden Gate cloning might be for you. This methodology relies on versatile Type IIS restriction enzymes. These cut outside their restriction sites allowing you to easily create customized overhangs to assemble up to 6 different fragments in a prescribed order. These fragments can be pooled in one tube and ligated in as little as 30 minutes!
Alternatively, you might consider Gibson cloning. In this technique, you first perform a round of PCR to generate DNA fragments with 15-40 bp of homology at their ends. These fragments are then combined with an exonuclease, a polymerase, and a DNA ligase to carry out a pooled set of ligations in an hour or less. The ligations combine the fragments in an order determined by their homology.
You might also consider FastCloning, first described by Chaokun Li and colleagues. Here you simply PCR-amplify your vector and your insert, DpnI digest away the templates, and transform the PCR products into E. coli. Ligases within the E. coli then fuse together the PCR products via overhangs left over from the PCR process (the precise mechanism isn’t well understood).
Finally, TOPO cloning is another good option to consider when time is of the essence. This method requires a vector backbone containing a topoisomerase I cut site, toposisomerase I, and a taq polymerase-generated insert. When mixed, topoisomerase I provides both the endonuclease and ligase function while 5’ adenosine overhangs left by Taq perfectly anneal with the topoisomerase I generated overhangs in the backbone. Once combined, the cut and ligation processes happen in a matter of minutes.
Cost:
Funding is usually limited - what cloning approach should you use if the budget is particularly tight? Most cloning processes will require a PCR reaction or two to generate templates for the cloning process. Time spent actually doing the experiments are also generally comparable so the best way to compare costs is to look at the prices of the additional enzymes required.
FastCloning may be the least expensive of all the options discussed as it only requires PCR and the inexpensive DpnI enzyme. Otherwise you let your E. coli do the work for you.
Traditional restriction enzyme and ligase based cloning is a relatively inexpensive way to get the job done, provided that you’ve already got the right enzymes laying around the lab. If not, you’ll be looking at about $50-$60 per enzyme. Once you’ve got a few of these common enzymes, however, your cost per cloning reaction drops dramatically, as one tube of enzyme should be enough for at least 50 cloning reactions. The ligase cost is comparable to that of the restriction enzymes and overall you’re looking at a price of a few dollars per reaction for traditional restriction cloning.
Sequence and Ligation Independent Cloning (SLIC) could also be a great choice, because the T4 polymerase needed for this method is relatively inexpensive and can be obtained for about $70, resulting in a price of a few dollars per reaction.
By comparison, Gibson assembly can cost over $15.00 per reaction if you purchase a pre-made mix. Nonetheless, Gibson assembly is a highly versatile and efficient way to generate a wide variety of constructs. Similar, Gateway cloning (discussed below) can run on the order of $20-30 per reaction but can be used to quickly generate multiple clones.
Difficulty:
If you’re an experienced cloner, you’re likely not intimidated by having to pick the right enzymes, optimize reagent concentration, or purify DNA fragments from gels. If you’re relatively new to the cloning game, however, or simply looking for a more streamlined approach, you might want to give Gateway a try. This method, developed by Invitrogen and based on lambda phage integration sequences, relies on site-specific recombination reactions catalyzed by BP Clonase and LR Clonase enzyme mixes. After generating the appropriate inserts by PCR, there is no need for restriction digests, gel purifications, or ligations. The general Gateway cloning protocol is fairly flexible and adaptable to cloning different types of DNA, such as cDNA, PCR fragments, or genomic DNA.
Also remember that commercially available cloning kits can often make the process much more streamlined, because they will come with comprehensive and tested protocols, as well as premixed and ready-to-use reagents. Thermo Fisher Scientific Inc., Origene, and NEB® are all good places to look for some helpful kits!
Efficiency:
Wouldn’t it be nice if after all the hard work involved with cloning, you would at least be guaranteed a successful result? While there are never any guarantees, some cloning techniques do tip the scales in your favor. For starters, Golden Gate cloning is extremely efficient due to the use of the versatile type II restriction enzymes. In Golden Gate cloning, the desired end construct does not contain the type IIS recognition sites, allowing you to keep the type IIS enzymes in the reaction to redigest vector reseals and other incorrect products. This feature biases the reaction in favor of the desired assembly to make the process close to 100% efficient.
Gateway cloning also offers a very high chance of success (>95% for a single insert), mostly due to the use of toxic ccdB to prevent any non-recombinant vectors from propagating. The ligation-independent methods like Gibson and SLIC are also quite efficient; although efficiency often declines as the number of fragments you’re hoping to assemble increases.
Multiple inserts:
There are some cloning methods that make working with multiple inserts a breeze. Among them are some of the homology based methods, like Gibson or SLIC, since one can pool the components and define their final order through sequence homology. Similarly, Golden Gate assembly allows one to simultaneously complete an ordered set of scarless ligations following digestion with Type II restriction enzymes. All of these options could be good choices if you have a particularly large number of inserts to assemble. You could also consider using Gateway if your insert number is a bit smaller, let’s say 4 or less.
The list goes on...
In this post, we’ve reviewed many of the most popular cloning methods out there, and I hope we’ve given you some tips for your next project. But keep in mind that there are many other cloning methods - including some that rely solely on PCR, and a host of commercially available cloning kits that one could choose from to perform these experiments. In most cases, there will be more than one right answer, so don’t be afraid to explore multiple approaches to find out which one works best for you. Happy cloning (and be sure to deposit your finished constructs at Addgene!).
References
1. Li, Chaokun, et al. "FastCloning: a highly simplified, purification-free, sequence-and ligation-independent PCR cloning method." BMC biotechnology 11.1 (2011): 92. PubMed PMID: 21992524. PubMed Central PMCID: PMC3207894.
Additional Resources on the Addgene Blog
- Read other Plasmids 101 blog posts
- New to plasmids? Read our article on What is a Plasmid?
- Download the Plasmids 101 eBook
Resources on Addgene.org0
- Check out protocols for plasmid cloning
- Find vector backbones
- Read our molecular biology reference
Topics: Plasmids 101, Plasmid Cloning, Plasmids







Leave a Comment