This post was contributed by guest blogger Meghan Monroy, a graduate student in Protein Science at the University of Connecticut.
Molecular cloning is the isolation, insertion and amplification of a recombinant DNA without sequence alteration. Molecular cloning techniques are some of the most widely used techniques in the biological sciences and constitute foundational elements of biomedical research. Traditional restriction cloning is one of the oldest of these techniques and is a multi-step process consisting of digestion, purification, ligation, and transformation. While restriction cloning is still routinely performed by many labs, a variety of other cloning techniques with higher efficiency and simpler procedures have been developed. Some of these include, TA cloning, ligation independent cloning, TOPO cloning, one step cloning, and overlap extension PCR. Although each type of cloning has its advantages, most scientists still encounter several struggles with these techniques: unwanted mutations due to excessive PCR cycles or low fidelity Taq DNA polymerase, the construction of specific sequences for base pair overhangs, insert and vector purification, and, most importantly, excessive time requirements. FastCloning is a simpler yet reliable cloning technique that was developed by Li, et al., in 2011. This method is ligation independent, it does not require purification of insert or vector products, nor does it require the use of specific sequences. Read on to learn how easy this process is and to get tips for applying it in your own lab.
Table 1: Comparison Between Standard Restriction Cloning and Fast Cloning
| Requirement | Restriction Cloning | Fast Cloning |
| Choose Restriction Enzymes | + | - |
| PCR Amplification | + | + |
| Digestion | + | + |
| Gel Purification | + | - |
| Ligation | + | - |
| Transformation | + | + |
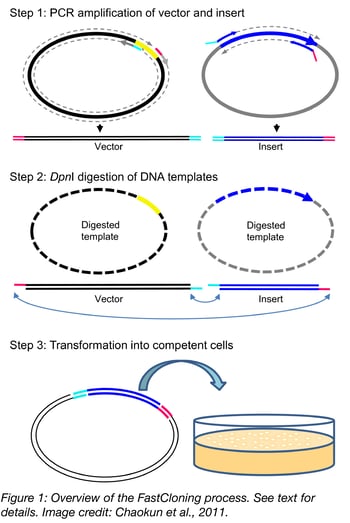 The FastCloning process in brief
The FastCloning process in brief
In FastCloning, you PCR both the vector and the insert in separate reactions and then combine the products and digest with methylation sensitive restriction enzyme DpnI to remove the templates. Digestion products are then transformed directly into chemically competent E. coli and transformants selected with the appropriate antibiotic. Although the precise mechanistic details of the process are not well understood, it is believed that the exonuclease activity of the polymerase generates sticky ends in the PCR reactions. These sticky ends are annealed and ligated in vivo.
Detailed FastCloning procedure
To start the FastCloning process, the insert and vector are first PCR amplified. The forward primer used to amplify the vector is located in the 3’ end of the multi-cloning site, and the reverse primer is located in the 5’ end of the multi-cloning site. The primers used to amplify the insert have sequences specific to the insert with an additional 15-17 bases that are complementary to the overlapping vector ends. See an overview of the PCR process and the resultant products of the reaction in the figure from Chaokun et al., 2011 to the right.
After PCR, you should confirm that the products were amplified by running a small aliquot of the reaction on an agarose gel. You should see bands with the appropriate sizes for your vector and insert.
Once you’ve confirmed that the PCR was successful, mix together the vector and insert PCR products with DpnI, and allow the digestion to take place for an hour at 37°C. This should be enough time to completely digest the template DNA. Note—templates must have been isolated from methylation competent E. coli for this procedure to work properly.
Directly transform chemically competent cells with the digested mixture. There is no need for further purification before transformation.
After transforming cells, you should perform colony PCR with vector specific primers to confirm that the products are the correct size (vector + insert). You should screen 3-4 colonies to ensure that you find the correct product. Once you find a colony with the appropriately sized product, purify and sequence the plasmid to confirm no mutations were introduced during PCR or any subsequent steps. Using high fidelity DNA polymerases, I have never had any mutations introduced into my inserts. However, with any PCR-based cloning technique there is a chance of introducing an unwanted mutation.
Tips for high efficiency cloning
Now that you have a better understanding of the step by step procedure of FastCloning, here are some tips for high efficiency cloning with this technique:
- Amplification efficiency is highest when using high fidelity DNA polymerases like phusion.
- An 18-cycle PCR leads to the most colonies after transformation.
- When a 1:1 vector:insert ratio is used in the DpnI digestion and 2 µL of digest product are used in the transformation, >70% of the colonies are correct.
Good luck with this fast and easy technique!
Many thanks to our guest blogger, Meghan Monroy.
 Meghan Monroy is currently an Outreach Data Curation Intern at Addgene. She is also a graduate student at the University of Connecticut interested in protein science.
Meghan Monroy is currently an Outreach Data Curation Intern at Addgene. She is also a graduate student at the University of Connecticut interested in protein science.
References:
1. Celie, Patrick HN, Annabel HA Parret, and Anastassis Perrakis. "Recombinant cloning strategies for protein expression." Current Opinion in Structural Biology 38 (2016): 145-154. PubMed PMID: 27391134.
2. Li, Chaokun, et al. "FastCloning: a highly simplified, purification-free, sequence-and ligation-independent PCR cloning method." BMC biotechnology 11.1 (2011): 1. PubMed PMID: 21992524. PubMed Central PMCID: PMC3207894.
Additional Resources on the Addgene Blog
- Plasmid Cloning by PCR
- Perform Site Directed Mutagenesis by PCR
- Catch up on Your Plasmid Background with our Plasmids 101 Posts
Resources on Addgene.org
- Find Empty Vectors for Your Research
- Find Additional Protocols
- Browse Our Special Collections for Plasmids from Your Field
Topics: Plasmid Cloning, Plasmids







Leave a Comment