Topoisomerase based cloning (TOPO cloning) is a DNA cloning method that does not use restriction enzymes or ligase, and requires no post-PCR procedures. Sounds easy right? The technique relies on the basic ability of complementary basepairs adenine (A) and thymine (T) to hybridize and form hydrogen bonds. This post focuses on "sticky end" TOPO (also called TOPO-TA) cloning; however, the TOPO cloning technique has also be adapted for blunt end cloning.
So how does TOPO cloning work?
As illustrated in the Figure below, the “A” overhang on the blue PCR product insert comes from using Taq polymerase for the amplification step since Taq polymerase leaves a single deoxyadenosine (A) at the 3' ends of PCR products. The complimentary “T” in the pair comes from a topoisomerase I-linearized backbone. DNA topoisomerase I (depicted as a green cloud) functions both as a restriction endonuclease and as a ligase by cleaving and rejoining supercoiled DNA ends to facilitate replication.
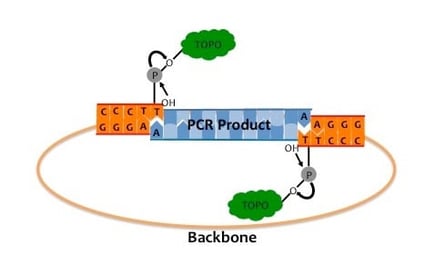 The TOPO technique specifically uses Vaccinia virus-isolated topoisomerase I as this enzyme recognizes the DNA sequence 5´-(C/T)CCTT-3' and digests double stranded DNA at this sequence. The energy from this breakage is stored as a covalent bond between the cleaved 3′ DNA strand and a tyrosyl residue of topoisomerase I (1). If a 5′ hydroxyl group from a different DNA strand comes along, it can attack this covalent bond thus joining the two DNA strands and releasing topoisomerase (2).
The TOPO technique specifically uses Vaccinia virus-isolated topoisomerase I as this enzyme recognizes the DNA sequence 5´-(C/T)CCTT-3' and digests double stranded DNA at this sequence. The energy from this breakage is stored as a covalent bond between the cleaved 3′ DNA strand and a tyrosyl residue of topoisomerase I (1). If a 5′ hydroxyl group from a different DNA strand comes along, it can attack this covalent bond thus joining the two DNA strands and releasing topoisomerase (2).
These days commercially available TOPO kits provide vectors or cloning arms with overhanging 3´ deoxythymidine (T) residues that are covalently linked to topoisomerase. Vectors in these kits also often have the topoisomerase site inserted into a beta-galactosidase cassette allowing a researcher to perform blue-white screening after transformation - self joining of the vector ends results in the production of blue colonies that do not need to be picked and sequenced for potential positive clones. Once you introduce your 3’-end “A” overhang insert, the magic of TOPO cloning happens.
Basic procedure
Let’s break down the steps needed for TOPO cloning:
1. Create Your PCR Product: Design standard primers (no need to add unique restriction sites on the ends) and amplify your sequence of interest with Taq polymerase using your favorite PCR protocol.
2. Set Up TOPO Cloning Reaction: Mix together the PCR product and TOPO Vector.
3. Incubate 5 Minutes at Room Temperature: You can place your reaction on ice if you are planning to transform right away OR you can store the reaction at -20C overnight.
4. Transform TOPO Cloning Reaction into Competent Cells: You can use your standard lab protocol for this; however, you should reduce your incubation time on ice to 5 minutes (incubating the full 30 minutes will not significantly improve the transformation efficiency).
5. Select and Analyze 10 White or Light Blue Colonies: You can confirm the presence of your insert by PCR, restriction digest, or sequencing.
Pro tips
- Do not add 5’ phosphates to your PCR primers; you need that free hydroxyl group!
- You may want to include extra extension time after the last cycle of PCR to make sure that the "A" gets added to all PCR products.
- Keep in mind that Taq polymerase has an error rate of about 1 in 3,500 bases. Typically polymerases with proofreading functionality are used in place of Taq to reduce error rates; however, proofreading polymerases will also remove all unpaired 3’ ends in your PCR product. If you need to decrease error rate, please use one of these methods to ensure your insert retains the 3' A overhang:
- Use a mixture of proofreading enzyme and Taq, with Taq used in an excess ratio of 10:1.
- Gel purify your PCR product and incubate it with buffer, Taq and dATPs at 72C for 10-15min.
- When mixing the PCR product with the TOPO vector, you may want to add extra salt to your reaction:
Topoisomerase I is released from the vector when the PCR product and vector ligate; however, it can potentially rebind and nick the newly ligated DNA. Salt helps prevent topoisomerase I from rebinding, which results in more intact molecules. (Note that the amount of salt you add will depend on whether you are planning on transforming your reaction into chemically or electro-competent E. coli - excess salt causes arcing during electroporation which would cause the electroporation to fail).
- When incubating at room temperature, it is not recommended that you exceed the 5 minute time limit (lower transformation efficiencies have been reported with longer incubation); however, you may need to incubate for 20-30 minutes if your PCR product is at a low concentration or you are cloning an extremely large insert.
- Since the standard ligation reaction is fairly quick, make sure you stay organized and prepare everything you need for the next step before proceeding.
- Pre-warming your antibiotic-containing plate prior to plating your transformation may allow you to see colonies within 8 hours.
References:
1. Shuman S. "Recombination mediated by vaccinia virus DNA topoisomerase I in Escherichia coli is sequence specific." Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10104-8. PubMed PMID: 1658796. PubMed Central PMCID: PMC52876.
2. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. Shuman S. J Biol Chem. 1994 Dec 23;269(51):32678-84. PubMed PMID: 7798275.
Additional Resources on the Addgene Blog
- Perform Site Directed Mutagenesis by PCR
- Use REPLACR Mutatgenesis to Easily Generate Insertion and Deletions in a Plasmids
- Learn How to Verify Your Plasmid
Resources on Addgene.org
- Find More Plasmid Cloning Protocols
- Catch Up On Your Basic Molecular Biology Techniques
- Watch Our Protocol Videos
Topics: Plasmids 101, Plasmid Cloning, Plasmids






Leave a Comment