This post was contributed by Shravanti Suresh from Iowa State University.
Since its appearance, SARS-CoV-2 has spread to almost every part of the world manifesting as a full-fledged pandemic. Containing the spread of this virus has become an utmost priority for countries around the world and to do so, the WHO recommends one strategy: testing, tracking, and social distancing.
With countries like South Korea, one of the earlier epicenters of the outbreak ultimately flattening the curve, it has become evident that widespread testing is crucial in controlling this pandemic. Currently, the CDC uses RT-qPCR tests to diagnose COVID-19 and some serological tests to determine past exposure. However, the limited availability of reagents and equipment and the long turnaround times, have led researchers to turn to other technologies like CRISPR.
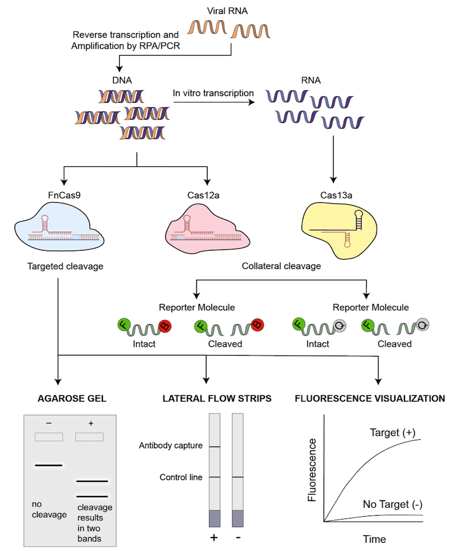 |
| Figure 1: An overview of the general schematic of CRISPR/Cas based COVID-19 testing methods. |
Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR based DETECTR assay
Sensitivity:10-100 copies of RNA per microliter
Time: ~40 minutes
Equipment required: 37°C heat block and 62°C heat block
Approach: Viral RNA is extracted and subject to isothermal amplification by primers that target the E and the N2 gene of SARS-CoV-2. If the Cas12a-sgRNA complex binds to the target sequences, Cas12a activity gets activated. This unleashes indiscriminate ssDNA cleavage activity which cleaves the supplied fluorescently labelled ssDNA substrates. Upon cleavage, the fluorescence is no longer quenched and can be visually detected using lateral flow strips. Since the probes are tagged with streptavidin, the lateral flow strip first captures the streptavidin portion of the probe via biotin. The fluorescent portion of the probe that is cut will continue to flow and be captured by an antibody further along on the strip.
Highlights: Of the 83 known COVID-positive samples tested, DETECTR has a positive prediction rate of 95% and a 100% negative prediction rate. It is also the fastest test that has been developed so far. A similar method has been optimized to detect the SARS-CoV-2 virus directly using saliva without having to perform RNA extraction but that may work only if there is a high enough (105 copies per microliter) viral load in the saliva (Lucia et al., 2020).
Detecting coronavirus using CRISPR-based SHERLOCK (Specific High sensitivity Enzymatic Reporter unLOCKing) technique
Sensitivity: Single molecule per milliliter can be detected – in the zeptomolar range
Time: ~1 hour
Equipment required: 37°C water bath and 42°C water bath
Approach: RNA is extracted from a sample and subject to isothermal recombinase polymerase amplification (RPA) using a commercially available amplification kit. In this case, regions of gene S and Orf1ab gene are amplified. RPA reactions also contain reverse transcriptase to produce cDNA. Isothermal amplification is done at one constant temperature, eliminating the need for thermocyclers. This is followed by in vitro transcription and Cas13 is then used to detect the amplified region of interest. Cas13 uses a complementary crRNA sequence to bind to the sample and upon binding, gets activated. This activation results in a trans cleavage reaction that cleaves a nearby RNA sequence that contains a fluorophore that is quenched when intact and released when cleaved. Cleavage results in fluorescence and this approach can be incorporated into a lateral-flow readout slip that allows rapid detection.
Highlights: Cas13a is not catalytically active in the presence of two or more mismatches in the cRNA:target duplex and will not give a positive result for highly similar viruses. Unlike Cas12a, Cas13a does not require strict sequence preferences at the target site, while Cas12a requires a PAM (Protospacer Adjacent Motif) for target cleavage. This test can be performed in two steps for challenging samples as in Two-Step-SHERLOCK or in a fast, high-throughput manner in one step as One-Pot-SHERLOCK. Two-step-SHERLOCK has a sensitivity in the zeptomolar range while One-step-SHERLOCK has a sensitivity in the femto-aptomolar range. For more details on the differences between these methods, check out this table.
All-in-one dual CRISPR/Cas12a (AIOD-CRISPR) assay – for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2
Sensitivity: 4.6-11 copies/microliter of RNA targets in a 40-minute incubation
Time: ~90 minutes
Equipment needed: 37°C heat block, LED blue light illuminator or UV light illuminator.
Approach: Highly similar to the SHERLOCK diagnostic test, this test varies in the sequence of steps and the Cas protein used. Here, all the components needed for the reaction are mixed in a single, one-pot reaction system and incubated at one temperature. Two Cas12a-crRNA complexes are designed, each binding to one strand of the reverse-transcribed target sequence that are present close to the primer recognition sites and are added separately. Using two crRNAs that bind to two regions improves sensitivity of the assay. This reaction also contains SSB, strand displacement DNA polymerase, recombinase, and the ssDNA-FQ reporters that exhibit fluorescence upon cleavage. Upon amplification by the RPA primers, the Cas12a-crRNA binds to the complementary region in the target sequence and this activity results in the trans cleavage of the ssDNA-FQ reporter resulting in fluorescence. Cas12a binds all the amplified target regions and results in the continuous cleavage of the ssDNA-FQ reporter. The resulting fluorescence can be observed under either no excitation light, UV light or LED blue light.
Highlights: This is a one-pot, one-temperature reaction, and is nearly single-molecule sensitive. In the case of HIV-1 and SARS-CoV-2 detection, AIOD-CRISPR without preamplification was able to detect as low as 1.2 copies DNA targets and 4.6 copies RNA targets in a 40 min incubation.
A scalable, easy-to-deploy, protocol for Cas13-based detection of SARS-CoV-2 genetic material (CREST)
Sensitivity: 10 copies of RNA per microliter
Time: Not mentioned in the paper, likely approximately 1-2 hours
Equipment needed: Mini PCR machines by DIY-Bio, P51 cardboard fluorescence visualizer with 9V battery
Approach :This preprint uses a previously developed method to detect SARS-CoV-2 in the samples. First, RNA is extracted and subject to reverse transcription. This is then amplified by PCR using Taq Polymerase and then transcribed to provide RNA substrates for Cas13. Cas13, upon finding complementarity between the gRNA and a given fluorescently labelled substrate, gets activated and performs non-specific collateral cleavage. This cleavage is coupled to fluorescence visualization. In this technique, however, instead of using lateral flow strips to detect the cleavage of fluorescently labelled RNA, the authors employ a P51 cardboard fluorescence visualizer that is powered by a 9V battery which uses a Blue LED and an orange filter to detect fluorescence upon cleavage.
Highlights: Reverse-transcribed RNA samples are amplified by PCR instead of RPA. Unlike RPA, this technique uses Taq polymerase which is widely available and less expensive. Instead of using the typical PCR machines, the authors use affordable, field-ready, Bluetooth-enabled PCR machines that can even be battery operated, made accessible by the DIY-Bio movement.
Rapid, field-deployable nucleobase detection and identification using FnCas9 (FELUDA)
Sensitivity: – 110 femtomolar
Time: Not mentioned, likely ~1-2 hours
Equipment needed: PCR machines/Heat blocks, Lateral flow strips, Agarose gel equipment, Fluorescent emission reader
Approach: FnCas9 Editor Linked Uniform Detection Assay (FELUDA) is a technique that uses FnCas9, a protein that is highly sensitive to the presence of mismatches within the DNA and the positions of these mismatches. To detect SARS-CoV-2 nucleic acids, the target sequence is amplified by PCR/RPA using biotinylated primers. These are then immobilized with streptavidin coated beads. Fluorescently labelled dCas9 complexes containing sgRNA that have no mismatches bind to the immobilised target sequences. Upon binding, changes in fluorescence can be visualised. Another approach was to apply the same concept to lateral flow strips that contain streptavidin which can bind to the biotinylated targets. They also tried to confirm target binding on agarose gels where a cleavage reaction upon proper binding results in two bands as opposed to a single band in the absence of cleavage.
Highlights: To detect non-PAM regions, PAM can be engineered into the primers. This method does not depend on collateral cleavage by the enzyme and instead relies on its activity on the target regions thus making it an affinity-based method. Due to the mismatch sensitivity of FnCas9, this is also a good way to diagnose single nucleotide variations. Also, FnCas9 cleaves in a range of temperatures varying between 10-50°C.
Visit Addgene's COVID-19 Plasmids and Resources

Many thanks to our guest bloggers Shravanti Surest from Iowa State University.
Shravanti Suresh is a PhD candidate who investigates cellular aspects of CRISPR/Cas immune systems in the Sashital Lab, Iowa State University. She is actively interested in science communication and enabling access to science in developing countries. You can follow her on Twitter @shra_krishna.
References
Azhar, M., Phutela, R., Ansari, A. H., Sinha, D., Sharma, N., Kumar, M., … Maiti, S. (2020). Rapid, field-deployable nucleobase detection and identification using FnCas9. https://doi.org/10.1101/2020.04.07.028167
Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan C-Y, Guevara H, Wadford DA, Chen JS, Chiu CY (2020) CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnology. https://doi.org/10.1038/s41587-020-0513-4
Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F (2019) SHERLOCK: nucleic acid detection with CRISPR nucleases. Nature Protocols 14:2986–3012 . https://doi.org/10.1038/s41596-019-0210-2
Lucia, C., Federico, P.-B., & Alejandra, G. C. (2020). An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. Cold Spring Harbor Laboratory. https://doi.org/10.1101/2020.02.29.971127
Rauch, J. N., Valois, E., Solley, S. C., Braig, F., Lach, R. S., Baxter, N. J., Kosik, K. S., Arias, C., Acosta-Alvear, D., & Wilson, M. Z. (2020). A Scalable, Easy-to-Deploy, Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material. Cold Spring Harbor Laboratory. https://doi.org/10.1101/2020.04.20.052159
Zhang, F., Abudayyeh, O. O. & Gootenberg, J. S. A protocol for detection of COVID-19 using CRISPR diagnostics. https://www.broadinstitute.org/files/publications/special/COVID-19%20detection%20(updated).pdf
Additional resources on the Addgene blog
- Read more about SHERLOCK and DETECTR
- Learn more about plasmids available for COVID-19 research
Resources on Addgene.org
- Get the basics about CRISPR in our CRISPR guide
- Browse our entire COVID-19 plasmid collection
Topics: CRISPR, Cas Proteins, COVID-19






Leave a Comment