Originally published Apr 30, 2020 and last updated Jul 12, 2021.
What started out as a small collection of plasmids for coronavirus research, including a handful of plasmids that have been in the repository for many years, has now grown into a collection of over 2,400 plasmids, with many available to scientists in industry. Since March 2020, we have received over 13,000 plasmids requests for COVID-19 related plasmids. These plasmids have reached labs in over 75 countries.
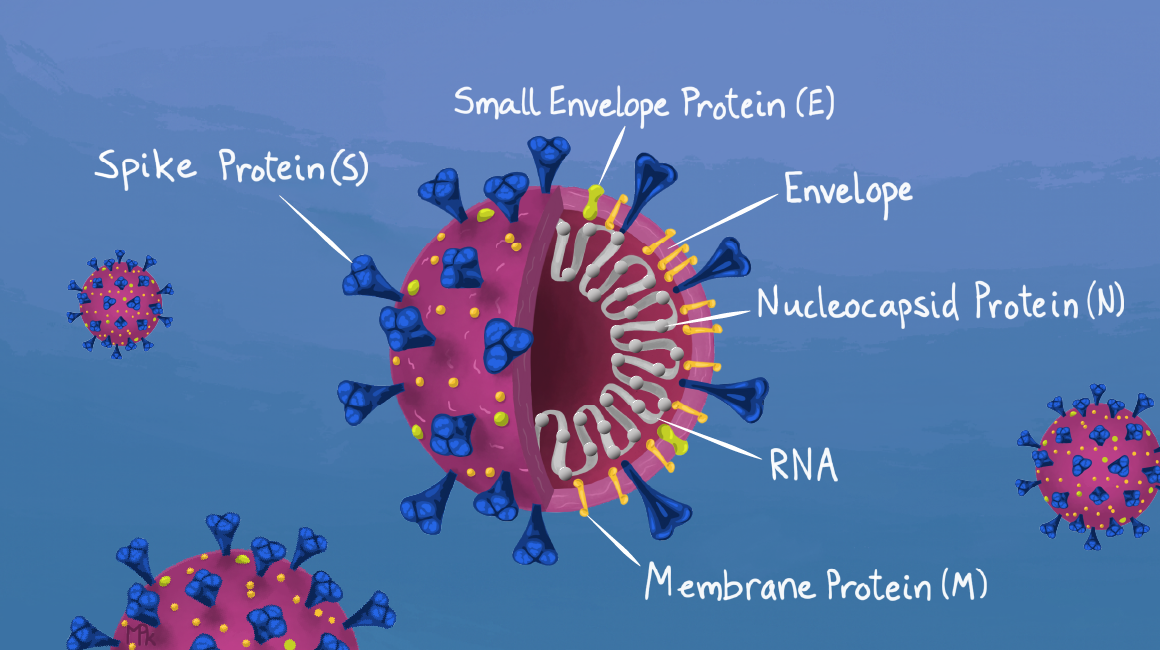 |
| Figure 1: SARS-CoV-2 proteins and other components. Image from Maya Kostman for the IGI. |
Here are some highlights of the plasmids shared over the last year and how these plasmids have been used to accelerate COVID-19 research in other labs.
Spike protein, variants, and pseudotyping
The SARS-CoV-2 virus depends on its spike protein to bind ACE2, the host cellular receptor. To more easily study the spike variants, scientists have developed ways to pseudotype the spike protein onto another virus. With the pseudovirus, scientists can study the mechanisms of infection and how to interfere with it in a BSL-2 lab instead of a BSL-3 lab. Jesse Bloom’s lab published a protocol and reagents for pseudotyping lentivirus with the SARS-CoV-2 spike protein. They deposited the plasmids used for this study at Addgene which include a C-terminal deletion in the spike protein to improve expression and pseudotyping efficiency. This plasmid was later used by another team of researchers for neutralization assays, part of a study that examined the repertoire of antibodies from 19 COVID-19 convalescent individuals (Tong et al., 2021).
Another version of the spike protein, called HexoPro, was developed by Jason McLellan’s lab. This version of the spike protein was developed based on 100 structure-guided spike designs and has improved protein yields and stability. This stabilized spike protein can be used for the development of vaccines and diagnostics (Hsieh et al., 2020).
As the virus spreads around the world, the spike protein has had many opportunities to evolve. Several variants are now circulating and plasmids containing these mutations have been deposited at Addgene. Plasmids deposited from Alejandro Balazs’s lab includes those for generating pseudovirus and those encoding different spike protein variants that can be incorporated into pseudoviruses. The lab used these pseudoviruses to determine how effective sera from vaccinated individuals are at neutralizing the different SARS-CoV-2 spike variants. You can learn more about their research in this article. To see other plasmids for SARS-CoV-2 spike pseudotyping, visit the SARS-CoV-2 Pseudotyped Virus collection page.
SARS-CoV-2 detection tools
There have been many detection tools developed over the last year for diagnosing COVID-19, aside from the widely used qPCR and antigen tests.
Labs including Jennifer Doudna’s lab and Feng Zheng’s lab had already developed nucleic acid detections tools based on Cas12a and Cas13 which could be repurposed for SARS-CoV-2 RNA detection. Early in the pandemic, labs around the world have been modifying these tools for COVID-19 detection with the ultimate goal to develop low cost and quick point-of-care diagnosis kits (reviewed in Tsang and LaManna, 2020). To see even more tools, head over to this blog post that highlights five different methods to use CRISPR to detect SARS-CoV-2.
Other protocols and tools for detection have been made openly available including those from Andrea Pauli and Julius Brennecke. They developed a method that uses loop-mediated isothermal amplification after reverse transcription (RT-LAMP) and a colorimetric readout that can be completed in just 45 minutes. The team deposited the reverse transcriptase, protease, and Bst polymerase constructs from their assay to Addgene.
Drew Endy’s and Philippa Marrack’s labs also deposited plasmids for producing COVID-19 diagnostic enzymes. Plasmids in this toolkit include ones for expressing MMLV reverse transcriptase and Taq polymerase that can be used in a one-step RT-qPCR reaction to detect SARS-CoV-2 nucleic acids.
Mammalian targets
During the 2003 SARS outbreak, Hyeryun Choe’s lab identified that ACE2 is the functional receptor for SARS-CoV. The lab also found that cathepsins play a role in SARS-CoV infection and deposited plasmids for the expression of cathepsin S, L, and B. This became a great starting point for SARS-CoV-2 researchers to investigate how the virus interacts with human cells. Addgene’s mammalian target plasmids express these and other targets such as TMPRSS2, FURIN, and more.
Recently, scientists used the genome-wide CRISPR libraries, Brunello (knock out) and Calabrese (activation), from Addgene to better understand which human genes are important for SARS-CoV-2 entry into human lung epithelial cells (Biering et al., 2021). In their search, they found mucins have an important role in viral entry, which are inhibitors of SARS-CoV-2 entry and become upregulated during viral infection.
Expression of SARS-CoV-2 viral proteins
In the early weeks of the COVID-19 pandemic, the Krogan lab at UCSF generated lentiviral vectors expressing SARS-CoV-2 open reading frames and identified interactions of these proteins with human proteins. Their deposited plasmids expressed 26 of the 29 SARS-CoV-2 proteins which all but one are cloned into pLVX vectors (for lentiviral packaging) and contain an IRES-Puro marker for stable cell clone selection. For more information on these plasmids and a behind-the-scenes glimpse of their work, head over to their blog post.
Months later, we partnered with Ginkgo Bioworks to provide synthesized SARS-CoV-2 expression plasmids available for many expression systems: bacterial, yeast, and mammalian. These plasmids contain both untagged and tagged versions of SARS-CoV-2 ORFs and include plasmids that are codon optimized. Browse the collection or learn more about it on the blog.
COVID-19 research is happening fast and we’re glad to join these efforts and help academic and industry scientists through plasmid sharing and more. As you’ve read above, some of these plasmids have been in the repository for many years while many were developed during the COVID-19 pandemic. Our mission to accelerate research through sharing reagents has never been so important.
References and resources
References
Biering SB, Sarnik SA, Wang E, Zengel JR, Sathyan V, Nguyenla X, Van Dis E, Catamura C, Yamashiro LH, Begeman A, Stark JC, Shon DJ, Fox DM, Puschnik AS, Bertozzi CR, Carette JE, Stanley SA, Harris E, Konermann S, Hsu PD (2021) Genome-wide, bidirectional CRISPR screens identify mucins as critical host factors modulating SARS-CoV-2 infection. bioRxiv. https://doi.org/10.1101/2021.04.22.440848
Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD (2020) Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 12:513. https://doi.org/10.3390/v12050513
Hsieh C-L, Goldsmith JA, Schaub JM, DiVenere AM, Kuo H-C, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, Chou C-W, Byrne PO, Hjorth CK, Johnson NV, Ludes-Meyers J, Nguyen AW, Park J, Wang N, Amengor D, Lavinder JJ, Ippolito GC, Maynard JA, Finkelstein IJ, McLellan JS (2020) Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369:1501–1505. https://doi.org/10.1126/science.abd0826
Tong P, Gautam A, Windsor I, Travers M, Chen Y, Garcia N, Whiteman MB, McKay LGA, Lelis FJN, Habibi S, Cai Y, Rennick LJ, Duprex WP, McCarthy KR, Lavine CL, Zuo T, Lin J, Zuiani A, Feldman J, MacDonald EA, Hauser BM, Griffths A, Seaman MS, Schmidt AG, Chen B, Neuberg D, Bajic G, Harrison SC, Wesemann DR (2021) Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike bioRxiv. https://doi.org/10.1101/2021.03.10.434840
Tsang J, LaManna CM (2020) Open Sharing During COVID-19: CRISPR-Based Detection Tools. The CRISPR Journal 3:142–145. https://doi.org/10.1089/crispr.2020.0030
Additional resources on the Addgene blog
- Learn more about mapping the SARS-CoV-2 - human protein-protein interaction
- Find tips for remote collaborations
- Browse all COVID-19 blog posts
Resources on Addgene.org
- Find COVID-19 resources on our COVID-19 and Coronavirus Plasmids & Resources page
- Stay up-to-date on Addgene's operations during the pandemic
- Find SARS-CoV-2 pseudovirus plasmids
Topics: Other Plasmid Tools, Plasmids, COVID-19






Leave a Comment