Originally published Aug 30, 2018 and updated April 16, 2020.
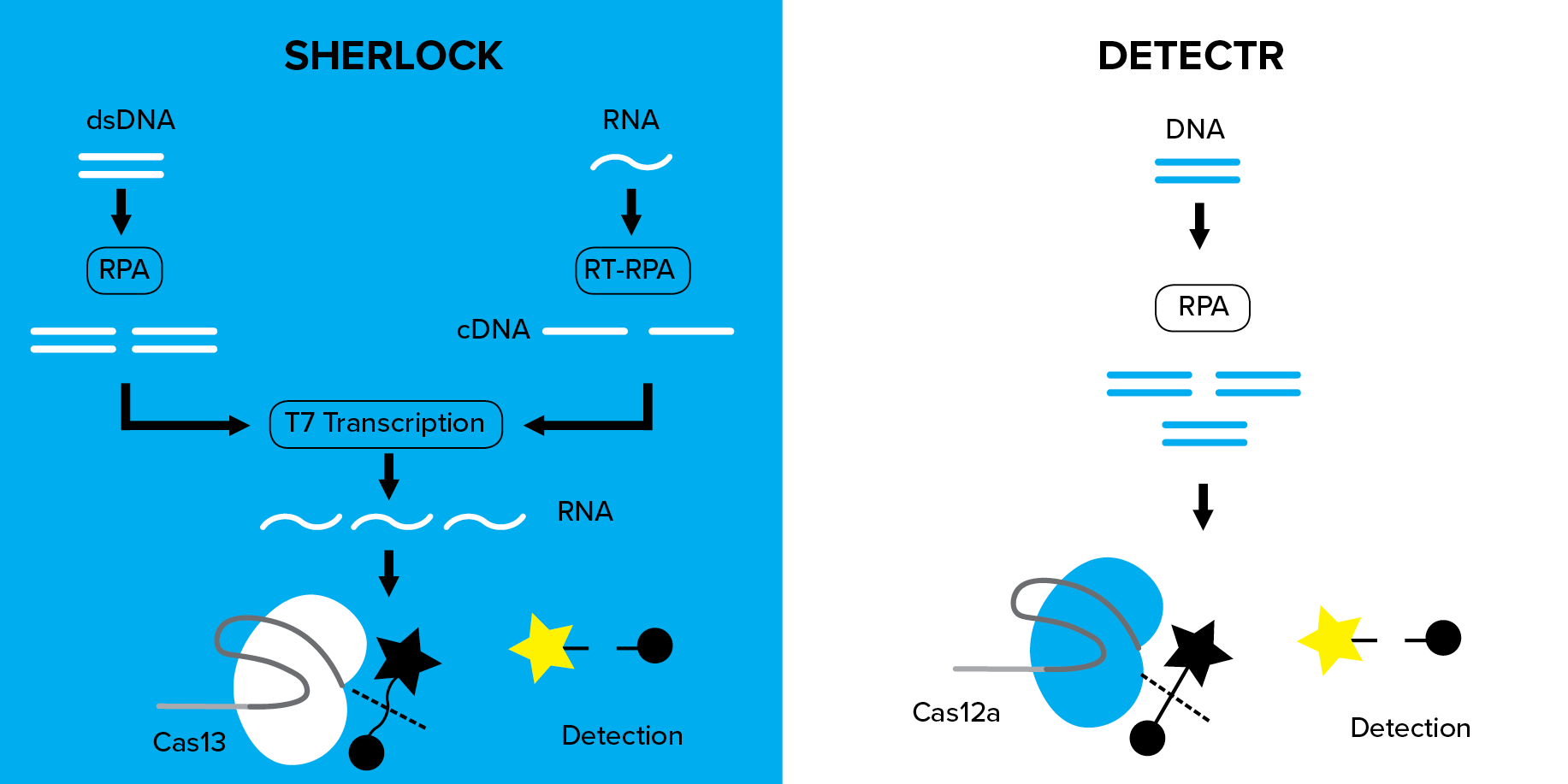 |
| Figure 1: Comparison of SHERLOCK and DETECTR nucleic acid detection methods. |
Sensitive and specific nucleic acid detection is crucial for clinical diagnostics, genotyping, and biotechnological advancements. Many methods of nucleic acid detection however, either lack the sensitivity or the specificity to detect nucleic acids at low concentrations and/or are too expensive, time-consuming, and complex to use outside of standard laboratories. In the case of the COVID-19 pandemic, qPCR can be used to diagnose the presence of SARS-CoV-2 RNA, but inadequate access to reagents and equipment has become a bottleneck.
In the last few years, scientists have utilized CRISPR-Cas9 protein variants, Cas13, and Cas12a, to develop simple, portable, and inexpensive platforms to reliably detect nucleic acids at the atomolar level.
The Zhang lab has adapted natural RNase activity of the Cas13 protein to develop and optimize the method termed Specific High Sensitivity Enzymatic Reporter UnLOCKING (SHERLOCK and SHERLOCKv2) (Gootenberg et al., 2017 and 2018). While the Doudna lab has used Cas12a’s non-specific ssDNA degradation to develop the method termed DNA Endonuclease Targeted CRISPR Trans Reporter (DETECTR) (Chen et al., 2018).
Both SHERLOCK and DETECTR harness the promiscuous cleavage and degradation of neighboring ssRNA and ssDNA by Cas13 and Cas12a, respectively, to cleave and activate a reporter. The detectable signal from this reporter can be measured and quantified to determine the presence and quantity of DNA, RNA or a mutation of interest. Together SHERLOCK and DETECTR demonstrate the power of CRISPR-based diagnostics.
SHERLOCK- Specific High Sensitivity Enzymatic Reporter UnLOCKING
Cas13 (C2c2) was first identified in 2016 by the Zhang lab as an RNA guided RNase (Abudayyeh et al 2016). Cas13 can be guided by a single CRISPR RNA (crRNA) to cleave ssRNA or mRNA. It also exhibits a “collateral effect” of non-specific ssRNA cleavage.
How does SHERLOCK work?
Cas13 can be programmed with crRNA to target an ssRNA of interest, for example, a sequence specific to a virus or pathogen. Once Cas13 recognizes and binds to the programmed sequence, it can promiscuously cleave surrounding ssRNA molecules. In SHERLOCK a quenched fluorescent ssRNA reporter is added to the reaction. Cleavage of the quenchable fluorescent RNA by the “activated” Cas13 produces a quantifiable signal that indicates the presence of your targeted nucleic acid. To increase the sensitivity of the assay, targeted DNA or RNA from a sample is first amplified using RPA (recombinase polymerase amplification) or reverse transcriptase (RT)-RPA, respectively. RPA is coupled with T7 transcription to convert amplified DNA to RNA for subsequent detection by Cas13. This amplification step in combination with the ssRNA reporter enables SHERLOCK to detect DNA or RNA with atomolar sensitivity and single base pair mismatch specificity.
SHERLOCKv2
SHERLOCK had a few limitations when it was first introduced in 2017 so the lab made further modifications to improve their detection system. They named this system SHERLOCKv2 for SHERLOCK version 2. Here are some of the improvements:
- SHERLOCKv2 uses far less primer in the pre-amplification step allowing for greater quantitation without compromising sensitivity.
- To detect multiple targets in one reaction, Cas enzymes, such as variations of Cas13 and Cas12a, are combined with different fluorescent reporters for each enzyme.
- In addition, Csm6, a CRISPR type-III effector nuclease, can be used in conjunction with Cas13 to amplify the signal of a single target. Csm6 can cleave ssRNA complementary to a crRNA of interest (Niewoehner and Jinek, 2016) and is conveniently activated by Cas13 ssRNA cleavage byproducts. Thus, binding of Cas13 to the programmed region of interest would lead to both the cleavage of a Cas13 specific reporter and the activation of Csm6. The Zhang lab showed that, by adding Csm6 and a Csm6 specific reporter (in the same fluorescent channel as the Cas13 reporter), they could significantly amplify the detection signal for a single target.
- SHERLOCKv2 was adapted so that a cleaved reporter could be detected on commercial lateral flow strips, similar to pregnancy tests. With lateral flow strips, the presence of your DNA or RNA of interest within a given sample is simply determined by the number of bands present on the strip. This type of readout allows for nucleic acid detection almost anywhere as lateral flow strips are easy to transport and work rapidly, providing reliable results in as little as an hour.
Find the SHERLOCK plasmids at Addgene!
Applications of the SHERLOCK detection system
The Zhang lab demonstrated that SHERLOCK could reliably distinguish between Zika and a closely-related virus, Dengue from multiple sample sources. SHERLOCK could also detect low-frequency cancer mutations from cell-free DNA fragments as well as health-related single nucleotide polymorphisms (SNPs) from human saliva.
SHERLOCKv2 provides a method to detect nucleic acids with high sensitivity and specificity without compromising speed, ease of use, and portability. This method can be used for an array of applications including clinical diagnostics (e.g. pathogen or virus detection), therapeutics and sensitive genotyping. The beauty of SHERLOCK systems is that it can be used easily and effectively in the lab and in the field (Myhrvold et al., 2018).
Now, the Zhang lab has shared a protocol for using SHERLOCK to detect SARS-CoV-2 RNA. The test is started using RNA purified from patient samples and can be read using a dipstick in under one hour. While the lab points out that the protocol is not approved for clinical use at this point, they hope that the protocol serves as a platform for advancing diagnostics.
DETECTR- DNA Endonuclease Targeted CRISPR Trans Reporter
Cas12a (Cpf1) is a CRISPR Cas variant that can also cleave dsDNA similarly to Cas9 (Zetsche et al., 2015 ). Cas12a however recognizes a different PAM site and generates 5’ and 3’ staggered ends after dsDNA breaks. The Doudna lab discovered Cas12a’s ability to cleave non-specific (trans) ssDNA and harnessed this ability to create a DNA detection platform called DETECTR.
How does DETECTR work?
DETECTR works similarly to SHERLOCK. Cas12a is targeted to a specific DNA sequence, such as the Human papilloma virus (HPV) genome, via a crRNA. An ssDNA-fluorescently quenched (FQ) reporter, which will produce a signal when the ssDNA is degraded, is added to the reaction. To enhance sensitivity, the DNA is first amplified through isothermal amplification by RPA. When Cas12a-cRNA base pairs with the dsDNA of interest, the DNase activity of Cas12a is initiated. Surrounding trans-ssDNA, including the ssDNA-FQ reporter are subsequently degraded at a rate of ~1,250 cuts per second. A quantifiable fluorescent signal designates the presence of your DNA of interest, in this case HPV.
As proof of concept, the lab demonstrated that DETECTR could accurately distinguish between two similar types of HPV, HPV16 and HPV18, from human cells, at atomolar levels, within one hour. DETECTR thus has the ability to rapidly detect nucleic acids with high selectivity and sensitivity from patient samples.
Find the DETECTR plasmids here!
Applications of DETECTR in diagnostics
DETECTR allows for simple and efficient detection of nucleic acids in a mixed population for an array of molecular and clinical diagnostic applications. The company Mammoth Biosciences has been started based on DETECTR technology with the mission to “provide a CRISPR-based platform on which an infinite number of tests” for biosensing can be built upon.
Mammoth Biosciences and UCSF recently adapted the DETECTR platform to detect SARS-CoV-2 using a lateral flow strip format. They published this in Nature Biotechnology. Like the SHERLOCK detection system, DETECTR has not been approved for diagnostics yet, but is undergoing validation studies.
Jennifer Tsang contributed to updating this article.
References
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:aaf5573 . https://doi.org/10.1126/science.aaf5573
Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan C-Y, Guevara H, Wadford DA, Chen JS, Chiu CY (2020) CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnology. https://doi.org/10.1038/s41587-020-0513-4
Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA (2018) CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360:436–439 . https://doi.org/10.1126/science.aar6245
Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356:438–442 . https://doi.org/10.1126/science.aam9321
Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F (2018) Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360:439–444 . https://doi.org/10.1126/science.aaq0179
Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA, Garcia KF, Barnes KG, Chak B, Mondini A, Nogueira ML, Isern S, Michael SF, Lorenzana I, Yozwiak NL, MacInnis BL, Bosch I, Gehrke L, Zhang F, Sabeti PC (2018) Field-deployable viral diagnostics using CRISPR-Cas13. Science 360:444–448 . https://doi.org/10.1126/science.aas8836
Niewoehner O, Jinek M (2016) Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 22:318–329 . https://doi.org/10.1261/rna.054098.115
Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV (2015) Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Molecular Cell 60:385–397 . https://doi.org/10.1016/j.molcel.2015.10.008
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163:759–771 . https://doi.org/10.1016/j.cell.2015.09.038
Additional resources on the Addgene Blog
- Learn about using toehold switches to detect Zika virus
- Learn more about targeting RNA with Cas13a (C2c2)
- Learn more about Cas12a (Cpf1) and its multiplex genome editing abilities
Resources on Addgene.org
- Checkout our CRISPR guide page
- Browse the CRISPR Collection
Topics: CRISPR, Cas Proteins, Other CRISPR Tools, COVID-19






Leave a Comment