 This post was contributed by guest blogger Jon Chow, an immunology PhD student at Harvard University.
This post was contributed by guest blogger Jon Chow, an immunology PhD student at Harvard University.
In my previous two posts, I’ve described the fundamentals of how to work with Drosophila as an experimental model organism. I then described the Gal4/UAS system used by geneticists to study gene function. In this final installment, I’ll provide a brief introduction as to how you can engineer new transgenic flies to study your favorite gene (YFG).
Sometimes, you want to use the Gal4/UAS system, but the available reagents do not match your needs. If YFG is thrillingly new, you might have to make a transgenic fly in order to study it. The process is fairly straightforward. Basically, you place your gene of interest in a plasmid and get the vector incorporated into the germline.
To begin this process, you will first need to do some molecular cloning. Depending on whether you are trying to overexpress or knockdown a gene, you might need different plasmids. Harvard’s DRSC/TRiP Functional Genomics Resource Center has various plasmids for overexpression and knockdown. Addgene also has a number of insect vectors to consider. An easy way to see what is available is to browse Addgene’s Collection under the insect expression system or by filtering by fly as species of gene.
Find Gal4 and UAS plasmids at Addgene
Choosing a vector to generate a transgenic fly
There are a few important points to consider when choosing the vector you’d like to use to generate your new fly line:
1. Where would you like the gene to be incorporated into the genome? Some vectors incorporate into the genome randomly, while others use attP sequence docking sites so that you can control exactly in which locus your transgene ends up (1):
- Random insertion has the advantage that you can get varying expression levels of your transgene in different fly lines. Also, if there is a chance you need to combine multiple transgenes in one fly, having the transgenes in different loci allows for genetic recombination so that the transgenes can be inherited on the same chromosome.
- Targeted Insertion allows for direct comparisons between lines expressing different variants of YFG because the transgenes are found in the same locus across fly lines (i.e. there won’t be positioning effects). When designing experiments where you want to express multiple transgenes, remember that you’ll need to integrate them all at different loci. Fortunately, there are many docking sites available for integration.
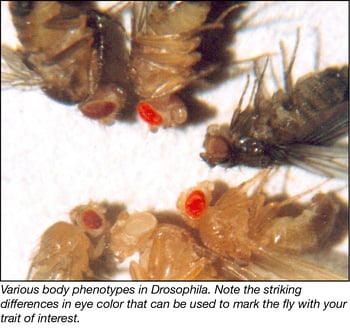 3. How would you like to select for your flies? Sure, the vector will have an antibiotic resistance gene to select for transformed bacteria, but you might be wondering, “How do I know which of my flies have the desired integration?” Not to worry! In flies, phenotypic selection markers make it easy to spot flies with the appropriate transgene. One such marker is the white gene, which restores eye color to fly lines where the endogenous white gene is knocked out. Flies with your integration will have eyes that are anywhere from light yellow to red. The variation in color depends on how transcriptionally active the genomic locus of integration is. The white gene is most often used, but other selection markers include vermillion (another gene controlling eye color) and yellow (a gene affecting the body color of the adult fly). This is important to note so that you know what to look for.
3. How would you like to select for your flies? Sure, the vector will have an antibiotic resistance gene to select for transformed bacteria, but you might be wondering, “How do I know which of my flies have the desired integration?” Not to worry! In flies, phenotypic selection markers make it easy to spot flies with the appropriate transgene. One such marker is the white gene, which restores eye color to fly lines where the endogenous white gene is knocked out. Flies with your integration will have eyes that are anywhere from light yellow to red. The variation in color depends on how transcriptionally active the genomic locus of integration is. The white gene is most often used, but other selection markers include vermillion (another gene controlling eye color) and yellow (a gene affecting the body color of the adult fly). This is important to note so that you know what to look for.
Making your transgenic fly and downstream experiments
Once your gene of interest is cloned into a plasmid, you can send your plasmid to a company. For a few hundred dollars, they will inject the plasmid into fly embryos and pick out flies where your transgene has been incorporated into the germline. Outsourcing the plasmid injections is much easier than doing it yourself. Depending on your strategy and the services offered by the company, you might have to map into which chromosome the transgene has integrated and make genetic crosses with balancer chromosomes to make genetically stable fly lines. After a few weeks of waiting, your flies will be ready for experimentation.
Drosophila are amenable to many different kinds of experimental assays depending on your interests. You can look at something as basic as survival. For those interested in development, organs of interest, such as the intestines, can be dissected at various times throughout the lifespan of the fly to determine the implications of your genetic manipulations. You can produce some very beautiful immunofluorescent images like in this paper (2). Metabolic assays such as Seahorse can be employed to look at oxygen consumption. Drosophila are also used in cancer research (3). The sky is the limit. As mentioned earlier, even genes not found in Drosophila can be studied. Using a Drosophila cell line, I’ve studied a transcription factor important for T cell differentiation (a cell type important for adaptive immune responses, which flies do not have!) (4). The power of Drosophila as a model organism is in its amenability to genetic manipulation.
This is just a primer so that you are aware of at least some of the possibilities. Basically, if you can devise an experimental assay that works with flies, you can utilize the resources of the Drosophila community to easily test your hypotheses.
Many thanks to our guest blogger Jon Chow.
 Jon Chow is finishing his immunology PhD at Harvard University. He has had nearly a decade of experience working with Drosophila as an experimental system. Jon enjoys studying the never-ending struggle between pathogens and their hosts. It’s not always clear which side he roots for. You can try to convince him to use Twitter @jonchowphd but he still reads “#” as a “pound symbol”.
Jon Chow is finishing his immunology PhD at Harvard University. He has had nearly a decade of experience working with Drosophila as an experimental system. Jon enjoys studying the never-ending struggle between pathogens and their hosts. It’s not always clear which side he roots for. You can try to convince him to use Twitter @jonchowphd but he still reads “#” as a “pound symbol”.
References
1. J. R. Bateman, et al. Site-Specific Transformation of Drosophila via PhiC31 Integrase-Mediated Cassette Exchange. Genetics. 173, 769–777 (2006). PubMed PMID: 16547094. PubMed Central PMCID: PMC1526508.
2. H. Jiang, et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 137, 1343–55 (2009). PubMed PMID: 19563763. PubMed Central PMCID: PMC2753793.
3. V. A. Rudrapatna, et al. Dev Dyn. 241. 107–118 (2012). PubMed PMID: 22038952. PubMed Central PMCID: PMC3677821.
4. J. R. Huh et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORγt activity. Nature. 472, 486–90 (2011). PubMed PMID: 21441909. PubMed Central PMCID: PMC3172133.
Additional Resources on the Addgene Blog
- Learn about Using Cre in Drosophila
- Read Other Genome Engineering Posts
- Use FLEx Vectors for Site-Specific Recombination
Resources at Addgene.org
- Browse Drosophila Plasmids
Topics: Drosophila, Other





Leave a Comment