There are approximately 2-4 million proteins per cubic micron in bacteria, yeast, and mammalian cells (Milo, 2013). The number of interactions between these proteins is hard to imagine yet alone study.
To elucidate these interactions scientists have long used antibody based affinity purification and mass spectrometry approaches. Recently however, scientists have developed newer methods termed proximity-based labeling to better capture these interactions in a vast array of biological contexts.
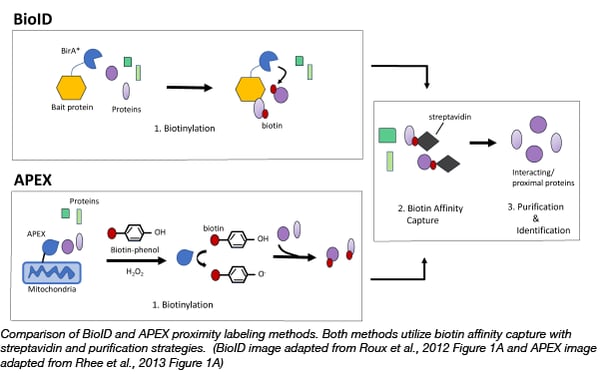
Proximity labelling relies on the labelling of neighboring proteins within a cell with biotin. Biotin, is a natural coenzyme, that strongly binds the glycoprotein, avidin or a similar protein, streptavidin. This strong interaction allows scientists to easily purify and identify any proteins that are tagged with biotin using avidin coated purification techniques. Due to the high affinity of biotin for avidin, proximity labelling can capture weak/transient protein interactions as well as be used for both soluble and insoluble proteins, which can be a challenge for classic antibody based affinity purification/mass spectrometry methods.
BioID: a promiscuous biotin ligase that can efficiently label neighboring proteins
The first biotin based proximity labelling technique, BioID, was developed in 2012 in the Brian Burke Lab (Roux et al., 2012). BioID utilizes the E. coli biotin ligase, BirA*, with a site mutation (R11G). This site mutation destabilizes the ligase, allowing for active biotin molecules to dissociate from the ligase and bind to exposed lysines on neighboring proteins. Thus to capture protein complexes, scientists can simply express a bait protein fused to BirA* in cells and then incubate those cells with exogenous biotin. After a few hours biotinylated proteins can be captured on streptavidin affinity beads or matrices. The bait protein can even target BirA* to specific subcellular locations, such as the nuclear envelope, allowing scientists to probe interactions in locations of interest.
Try BioID for your proximity labeling experiment!
APEX: engineered ascorbate peroxidase that can rapidly biotin label proteins
A year after BioID became available, the Alice Ting Lab demonstrated that engineered ascorbate peroxidase (APEX), used as a genetic tag for electron microscopy, could be used for efficient proximity labeling (Rhee et al., 2013). APEX catalyzes the oxidation of biotin-phenol to the short-lived biotin-phenoxyl radical in the presence of hydrogen peroxide. This radical reacts with electron-rich amino acids such as tyrosine on neighboring proteins resulting in their biotinylation. The advantage of APEX is its speed, labeling neighboring proteins in minutes rather than hours like BioID. APEX has allowed scientists to identify the proteomic map of the human mitochondrial matrix (Rhee et al., 2013) and determine the topology of the mitochondrial calcium uniporter (Martell et al., 2012). It is important to note that the biotin-phenol reagent can be toxic to living cells thus APEX is not suitable for organoid or in vivo studies (Che and Khavari, 2017).
Find APEX plasmids at Addgene!
 Since the development of BioID and APEX, optimized versions of BioID and APEX have been developed as well as new proximity labelling techniques. These newer tools are also currently available at Addgene.
Since the development of BioID and APEX, optimized versions of BioID and APEX have been developed as well as new proximity labelling techniques. These newer tools are also currently available at Addgene.
BioID2: A smaller improved promiscuous biotin ligase
The Roux Lab in 2016 optimized BioID to create BioID2, a smaller and improved promiscuous biotin ligase (Kim et al 2016). BioID2 consists of a humanized biotin ligase from A. aeolicus, which lacks the DNA-binding domain (233 amino acids) found in the E. coli version. It also contains a single mutation (R40G) in a conserved residue in the biotin catalytic domain to allow for promiscuous biotinylation. Compared to BioID, BioID2 exhibits enhanced proximal labelling, requires less biotin, and allows scientists to more selectively target fusion proteins to specific locations in the cell.
APEX2: A more sensitive ascorbate peroxidase for proximity labeling
A major limitation of APEX was its low sensitivity. Sensitivity could be increased with higher expression levels but for many fusion proteins this would lead to protein/organelle aggregation. The Ting lab hypothesized that the low sensitivity was due to suboptimal folding or stability. Thus they conducted multiple rounds of directed evolution until they identified a mutant with significantly increased sensitivity that did not affect the function or efficiency of the original APEX (Lam et al. 2014). They named this mutant APEX2.
TurboID and minturbo: Rapid and non-toxic proximity labeling
TurboID and miniTurbo were developed in the Ting Lab in 2018 to increase the speed of proximity labeling by promiscuous biotin ligases (Branon et al., 2018). BioID and BioID2 can take hours to label proteins and APEX2, although it can rapidly label proteins, requires H2O2 which can be toxic to living cells. To circumvent these issues, the Ting lab utilized yeast display-based directed evolution to generate two mutants, the 35 kD TurboID, which has 15 mutations compared to BirA and the 28kD miniTurbo, which has the N-terminus domain deleted and 13 mutations compared to wild type BirA. TurboID and miniTurbo both enable biotin labeling in 10 minutes without any issues in toxicity. TurboID’s signal is significantly greater than miniTurbo at earlier time points but miniTurbo exhibits less labeling prior to the addition of exogenous biotin allowing for more precise temporal control.
Find plasmids for TurboID and miniturbo
Split-BioID: A conditional proteomics approach to monitor protein-protein interactions
Split-BioID combines a protein fragment complementation (PCA) technique with proximity labelling to validate binary protein-protein interaction as well as identify additional interacting proteins in one simple experiment. PCA is an in vivo technique where two proteins are fused to complementary reporter proteins that can only assemble when in close proximity. Split-BioID consists of a split BirA* molecule that can reassemble to restore activity when fused to two interacting proteins (Schopp et al., 2016). Thus in one experiment a scientist can confirm and monitor the interaction of two proteins while also biotin labeling and purifying additional proteins from a complex of interest. For more information, read the Split-BioID blog post.
RaPID: RNA-protein interaction detection
So far, we’ve covered ways to study protein-protein interactions, but scientists have also developed ways to detect RNA-protein interactions that can regulate multiple biological functions. RapID (RNA-protein interaction detection) uses proximity protein labeling based on BioID to identify RNA-protein interactions (Ramanathan et al., 2018). To develop RapID, scientists engineered a mutant BirA* from B. subtilis, termed BASU. BASU exhibits 1,000 fold faster kinetics and increases the signal to noise ratio 30-fold compared to the standard E. coli BirA* mutant. RapID consists of this mutant BirA* and an RNA component that contains an RNA motif of interest. Any proteins that bind the RNA motif will be biotinylated. Read more about RapID in this blog!
EXCELL: Proximity labeling for the detection of cell-cell interactions
EXCELL is an enzyme-mediated proximity cell labeling strategy to detect cell-cell interactions in vivo (Ge et al., 2019). This technique relies on an S. auerus transpeptidase sortase A variant (mgSrtA), which is capable of promiscuously labeling various cell surface proteins that contain a monoglycine residue at the N terminus with a small peptide (LPETG). In EXCELL, the mgSrtA variant can be displayed on the surface of a cell of interest, where it can label interacting cell surface proteins with a biotin tagged LPETG peptide. Biotin tagged proteins can then be purified using streptavidin and separated via flow cytometry.
Find plasmids for EXCELL here!
Since BioID was developed in 2012, proximity labelling has become a powerful technique for studying protein-protein interactions, RNA-protein interactions, and even more recently cell-cell interactions. If you’re looking to identify interactions with your favorite protein or RNA motif, give these methods a try.
References:
Branon, Tess C., et al. "Efficient proximity labeling in living cells and organisms with TurboID." Nature biotechnology 36.9 (2018): 880. PubMed PMID: 30125270. PubMed Central PMCID: PMC6126969.
Che, Yonglu, and Paul A. Khavari. "Research Techniques Made Simple: Emerging Methods to Elucidate Protein Interactions through Spatial Proximity." Journal of Investigative Dermatology 137.12 (2017): e197-e203. PubMed PMID: 29169465.
Ge, Yun, et al. "Enzyme-Mediated Intercellular Proximity Labeling for Detecting Cell–Cell Interactions." Journal of the American Chemical Society 141.5 (2019): 1833-1837. PubMed PMID: 30676735.
Kim, D. I., Jensen, S. C., Noble, K. A., KC, B., Roux, K. H., Motamedchaboki, K., & Roux, K. J. (2016). An improved smaller biotin ligase for BioID proximity labeling. Molecular biology of the cell, 27(8), 1188-1196. PubMed PMID: 26912792. PubMed Central PMCID: PMC4831873.
Lam, Stephanie S., et al. "Directed evolution of APEX2 for electron microscopy and proximity labeling." Nature methods 12.1 (2015): 51. PubMed PMID: 25419960. PubMed Central PMCID: PMC4296904.
Martell, Jeffrey D., et al. "Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy." Nature biotechnology 30.11 (2012): 1143. PubMed PMID: 23086203. PubMed Central PMCID: PMC3699407.
Milo, Ron. "What is the total number of protein molecules per cell volume? A call to rethink some published values." Bioessays 35.12 (2013): 1050-1055. PubMed PMID: 24114984. PubMed Central PMCID: PMC3910158.
Ramanathan, Muthukumar, et al. "RNA–protein interaction detection in living cells." Nature methods 15.3 (2018): 207. PubMed PMID: 29400715. PubMed Central PMCID: PMC5886736.
Roux, Kyle J., et al. "A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells." J Cell Biol 196.6 (2012): 801-810. PubMed PMID: 22412018. PubMed Central PMCID: PMC3308701.
Rhee, Hyun-Woo, et al. "Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging." Science 339.6125 (2013): 1328-1331. PubMed PMID: 23371551. PubMed Central PMCID: PMC3916822.
Schopp, Isabel Myriam, et al. "Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes." Nature communications 8 (2017): 15690. PubMed PMID: 28585547. PubMed Central PMCID: PMC5467174.
Additional resources on the Addgene blog
Other resourcesTopics: Plasmid Tags, Plasmids






Leave a Comment