The central dogma in molecular biology is DNA→RNA→Protein. To synthesize a particular protein DNA must first be transcribed into messenger RNA (mRNA). mRNA can then be translated at the ribosome into polypeptide chains that make up the primary structure of proteins. Most proteins are then modified via an array of post-translational modifications including protein folding, formation of disulfide bridges, glycosylation and acetylation to create functional, stable proteins. Protein expression refers to the second step of this process: the synthesis of proteins from mRNA and the addition of post-translational modifications
Learn more molecular biology in our Plasmids 101 eBook!
Researchers use various techniques to control protein expression for experimental, biotechnological, and medical applications. Researches can visualize proteins in vivo by tagging them with fluorescent proteins to study localization or purify proteins to study their structure, interactions and functions. Proteins can also be purified for use in molecular biology research (eg. polymerases and other enzymes might be purified and used to manipulate DNA), or in medicine (e.g. insulin).
Proteins, unlike DNA which can be relatively easily synthesized, must be produced using complex mixtures derived from cells or using live cells. There are several types of expression systems used for protein production and purification. These include mammalian, insect, bacterial, plant, yeast and cell free expression systems.
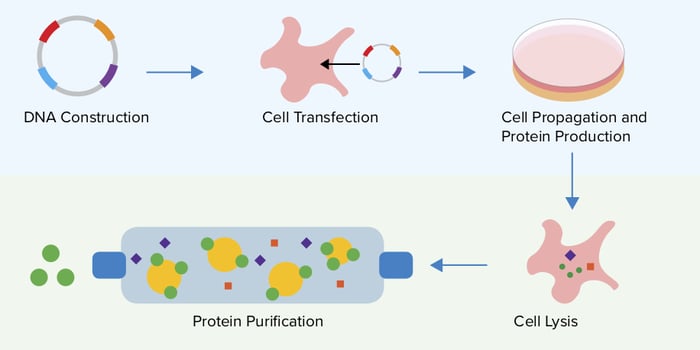
Overall the general strategy for protein expression consists of transfecting cells with your DNA template of choice and allowing these cells to transcribe, translate and modify your protein of interest. Modified proteins can then be extracted from lysed cells through the use of protein tags and separated from contaminants using a variety of purification methods. Deciding which expression system to use depends on several factors:
- The protein you are trying to express
- How much protein you need
- Your plans for downstream applications
In this blog post we will summarize some of the more common expression systems including their advantages and caveats to keep in mind before choosing a system.
Mammalian expression systems
Mammalian cells are an ideal system for the expression of mammalian proteins that require multiple post-translation modifications for proper protein function. Most DNA constructs designed for mammalian expression utilize viral promoters (SV40, CMV, and RSV) for robust expression post-transfection. Mammalian systems can express proteins both transiently or through stable cell lines. Both methods produce high protein yields if transfection is successful.
Some mammalian systems also allow for control over when a protein is expressed through the use of constitutive and inducible promoters. Inducible promoters are extremely useful if a desired protein product is toxic to cells at high concentrations. Despite their advantages, mammalian expressions systems do require demanding cell culture conditions compared to other systems.
Insect expression systems
Insect cells can also be used to produce complex eukaryotic proteins with the correct post-translational modifications. There are two types of insect expression systems; baculovirus infected and non-lytic insect cells.
Baculovirus expression systems are very powerful for high level recombinant protein expression. These systems enable high expression of very complex, glycosylated proteins that cannot be produced in E. coli or yeast cells. The only problem with baculovirus systems is that the infected host cell is eventually lysed. Cell lysis halts protein production, but there are non-lytic insect cell expression systems (sf9, Sf21, Hi-5 cells) that allow for continuous expression of genes integrated into the insect cell genome. Both of these types of insect expression systems can be scaled up for production of large amounts of protein.
Some drawbacks of insect cell expression systems include that virus production can be quite time consuming and that insect cells require demanding culture conditions similar to mammalian expression systems.
Bacterial expression systems
When one wants to produce vast quantities of protein rapidly and cheaply, a bacterial host cell is almost always the answer. E. coli is definitely one of the most popular hosts for protein expression with several strains that are specialized for protein expression. Protein expression in bacteria is quite simple; DNA coding for your protein of interest is inserted into a plasmid expression vector that is then transformed into a bacterial cell. Transformed cells propagate, are induced to produce your protein of interest, and then lysed. Protein can then be purified from the cellular debris.
There are several popular DNA vectors that can be used to produce large amounts of protein in bacterial cells: the pET, pRSET, Gateway pDEST, and pBAD vectors for example. Protein expression from each of these vectors is controlled by a different promoter resulting in different levels of expression from each vector; lower expression may be required if your protein is toxic to E. coli. Of all the vectors, pET, under the control of the T7 lac promoter and induced by lactose, provides the highest level of protein expression.
Despite their ease of use, it is important to note that bacteria usually cannot produce functional multi-domain mammalian proteins as bacterial cells are not equipped to add appropriate post-translational modifications. In addition many proteins produced by bacteria become insoluble, forming inclusion bodies that are difficult to extract without harsh reagents and patience.
Plant expression systems
Plants provide a cheap and low-tech means of mass expression of recombinant proteins. Many cells from various types of plants such as maize, tobacco, rice, sugarcane, and even tubers of potatoes have been used for protein expression.
Plant systems share many of the same features and processing requirements as mammalian cell expression systems, including the majority of complex post-translational modifications. Extraction and purification of recombinant proteins from plants however can be expensive and time consuming as plant tissues themselves are biochemically complex.
To circumvent these issues, scientists have taken advantage of the natural secretion of biochemicals and proteins through plant roots. Tagging recombinant proteins with a naturally secreted plant peptide allows for easier access and purification of a desired protein. Despite being a rather nascent technology, plant cells have been used to express a wide range of proteins including antibodies and pharmaceuticals, specifically interleukins.
Yeast expression systems
Yeast are a great expression system to generate large quantities of recombinant eukaryotic proteins. Although many species of yeast can be used for protein expression, S. cerevisiae, is the most reliable and frequently used species due to its use as a model organism in genetics and biochemistry.
When using S. cerevisiae, researchers often place recombinant proteins under the control of the galactose inducible promoter (GAL). Other commonly used promoters include the phosphate and copper inducible PHO5 and CUP1 promoters respectively. Yeast cells are grown in well-defined media and can be easily adapted to fermentation allowing for large-scale, stable production of proteins.
In general yeast expression systems are easier and cheaper to work with than mammalian cells, and are often capable of modifying complex proteins unlike bacterial systems. Yeast cells, however, have a slower growth rate than bacterial cells and growing conditions often need to be optimized. Yeast cells are also known for hyperglycosylating proteins which may be an issue depending on your protein of choice.
Cell free expression systems
In cell-free expression systems, proteins are assembled in vitro using purified components of the transcription and translation machinery. These include ribosomes, RNA polymerase, tRNAs, ribonucleotides and amino acids. Cell-free expression systems are ideal for fast assembly of more than one protein in one reaction. A major advantage of these systems system is their ability to assemble proteins with labelled or modified amino acids that are useful in different downstream applications. Cell-free expression systems however, are expensive and very technically challenging to use.
 Alyssa D. Cecchetelli is a Scientist at Addgene. She received her PhD from Northeastern University and is particularly interested in cell signaling and communication. She loves being able to help the scientific community share plasmids.
Alyssa D. Cecchetelli is a Scientist at Addgene. She received her PhD from Northeastern University and is particularly interested in cell signaling and communication. She loves being able to help the scientific community share plasmids.
Additional Resources
- Thermofisher Protein Expression Systems
- Recombinant protein expression in Escherichia coli: advances and challenges
- Production of recombinant proteins in plant root exudates
Resources on the Addgene Blog
- Learn about inducible promoters
- Learn about repressible promoters
- Check out protein tags
Resources on Addgene.org
Topics: Plasmids 101, Plasmids






Leave a Comment