Sometimes it feels like DNA and protein get all the attention.There are numerous ways to detect DNA-protein interactions or to analyze chromatin states (CHIP-seq, FAIRE-seq, Cut & Run) and to detect protein-protein interactions (yeast-two hybrid, Co-IP, BioID), and that’s just to name a few. But what if you want to study RNA-protein interactions? The characterization of RNA-protein interactions has lagged behind, likely due to limitations of current means to detect RNA-protein interactions. To address this need, the Khavari lab at Stanford created the RNA-protein interaction detection (RaPID) method. RaPID borrows the E. coli biotin ligase BirA* from BioID and allows a researcher to identify proteins that bind an RNA motif of interest in living cells.
Challenges with existing RNA-protein interaction methods
- Not in a native context. While native purification preserves RNA structure and naturally occurring complexes present in the cell, it can also introduce non-physiological RNA-protein interactions due to sample processing.
- Require prior knowledge that the protein of interest interacts with RNA. Methods like cross-linking immunoprecipitation (CLIP) identify RNA-protein interactions by cross-linking RNAs and proteins with UV light, followed by an antibody pulldown of a protein of interest, much like using ChIP for studying protein and DNA interactions. Unfortunately, using proteins as bait has limited utility for identifying novel RNA-binding proteins that interact with an RNA of interest.
- Current labeling methods are not ideal for in vivo work. BirA* requires 16-18 hours to sufficiently biotinylate interacting proteins, which results in poor spatiotemporal resolution for in vivo work. APEX2, a faster peroxidase-based labeling approach, is an alternative option but it’s toxic to tissues and model organisms.
Detecting RNA-protein interactions with RaPID
RaPID is similar to the BioID platform for identifying protein-protein interactions, but RaPID identifies RNA-protein interactions. Both methods rely on the promiscuous E. coli biotin ligase, BirA*, to biotinylate proteins that are within ~10 nm of the bait, which for RaPID is an RNA motif of interest. See figure 1 below for details on the key components of RaPID.
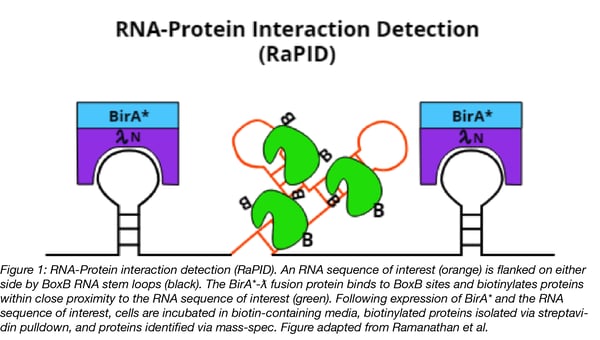
Ramanathan et al. also tweaked BirA* to increase its rate of biotinylation. The improved enzyme, called BASU, is derived from a BirA* homolog found in B. subtilis. BASU was compared head-to-head with the E. coli and BioID2 BirA*s in the context of RaPID for their ability to detect a known RNA-protein interaction. In as little was one minute BASU generated a strong streptavidin signal (~250-fold enrichment) when compared to a scrambled RNA motif control). BioID2 had no detectable signal at the same time point. E. coli’s BirA* was tested at its optimum 18 hour time point, but one minute of labeling with BASU still generated a >30-fold enrichment compared to E. coli’s BirA*. Faster labeling kinetics also means that BASU could potentially be used in vivo to study the the role played by RNA-protein interactions in events that occur on a short timescale, such as during drug action or cell signaling.
Limitations of RaPID
Like other BirA*-based proximity labeling methods, RaPID cannot distinguish between proteins that directly or indirectly interact with the bait. Also, RaPID can’t be used to study endogenous RNAs at their physiological expression levels; however, knock-in of BoxB sites into endogenous DNA sequences could allow one to study RNAs expressed from their native regulatory elements.
Controls for RaPID experiments
RaPID negative controls include both scrambled RNA motifs and cells transfected with BirA* protein alone (i.e. no RNA bait). Ramanathan et al. also used the Contamination Repository for Affinity Purification (CRAPome), to identify highly biotinylated background proteins reported by other research groups that performed BirA*-based affinity purification mass spec experiments. These background proteins were then subtracted from Ramanathan et al.’s RaPID datasets.
Applications of RaPID
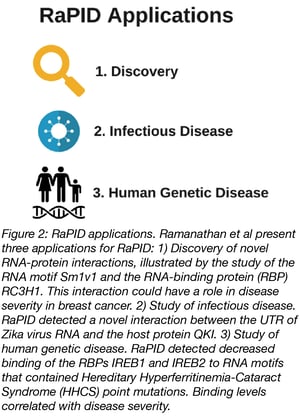
RaPID sounds like a nifty technique, but what kinds of questions can it help answer? Ramanathan et al. outline three applications of RaPID:
- Discovery. To discover novel RNA-protein interactions, Ramanathan et al. designed an RNA bait containing three well conserved but not well characterized RNA motifs. This bait identified a new interaction: binding of the RC2H1 protein to the SM1v1 RNA motif. It’s already known that RC3H1 and its paralog RC2H2 are RNA binding proteins that promote RNA decay and their upregulation is correlated with poor survival in breast cancer patients. By digging into the literature and analyzing data from The Cancer Genome Atlas, Ramanathan et al. found evidence that suggests changes in RC2H1 protein expression and post-transcriptional regulation of SM1v1-containing RNAs could play a role in the severity of breast cancer.
- Infectious Disease. RaPID can also help identify which host proteins interact with viral RNAs. RNA viruses, like Zika, often trick host proteins into binding their untranslated regions (UTRs) to aid in the viral life cycle. Ramanathan et al. used RaPID to see what host proteins interact with Zika’s UTR and identified QKI, an RNA-binding protein that’s highly expressed in neural progenitor cells. Knock-down of QKI decreased Zika viral RNA levels by 90%, suggesting that QKI is a host protein critical to Zika replication.
- Human genetic disease. RaPID can also be used to test how RNA variants associated with human disease affect protein binding patterns. Hereditary Hyperferritinemia-Cataract Syndrome (HHCS), a disease characterized by increased iron serum levels and early-onset cataracts, is characterized by point mutations in the Iron Response Element (IRE) RNA motif of the ferritin light chain RNA transcript. IRE motifs are bound by IRE binding proteins, IREB1 and IREB2, which post-transcriptionally regulate iron metabolism pathway transcripts. By using RaPID, Ramanathan et al. found that all HHCS-associated sequences had lower IREB2 binding enrichment compared to wild-type sequences and that loss of binding correlated with increased serum iron levels seen in HHCS patients.
These are just three examples of how RaPID can be used to study RNA-protein interactions. Do you have a question that RaPID could help answer? The RaPID plasmids are available from Addgene!
References
Ramanathan, M., Majzoub, K., Rao, D.S., Neela, P.H., Zarnegar, B.J., Mondal, S., Roth, J.G., Gai, H., Kovalski, J.R., Siprashvili, Z., Palmer, T.D., Carette, J.E., & Khavari, P.A. (2018). RNA–protein interaction detection in living cells. Nature Methods, 15, 207-212. PubMed PMID: 29400715. PubMed Central PMCID: PMC5886736.
McHugh, C.A., Russell, P., & Guttman, M. (2014). Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biology. PubMed PMID: 24467948 PubMed Central PMCID: PMC4054858.
Additional Resources on the Addgene Blog
- Study Protein-DNA Interactions with CUT&RUN
- Study Protein-Protein Interactions with Split-BioID
- Find resources for studying chromatin
Resources on Addgene.org
- Find Split-BioID plasmids
- Find CRISPR plasmids for labeling genomic regions
- Find CRISPR plasmids for purifying specific genomic regions
Topics: Other Plasmid Tools, Plasmids





Leave a Comment