The PAM… that sneaky little bit of sequence that you hope is present next to the "perfect" guide sequence for your genome engineering experiment. With CRISPR entering the clinic for correction of disease-causing alleles, and the growing need for gene editing in research, the old PAM constraints just won’t cut it anymore. Say hello to PAM-free and PAM-flexible nucleases! In this blog, we will review Cas enzymes, which have a flexible or non-existent PAM requirement, and how these proteins are advantageous in today’s genome editing landscape.
The need for PAM-less editing
Guides for CRISPR are typically selected by the ‘quality’ of the gRNA sequence, including propensity for off-targets, secondary structures, etc., as well as the presence of a Protospacer Adjacent Motif (PAM) sequence. If the gRNA sequence is suitable, but there is no PAM next to the desired cut site, then a further-away gRNA where there is an available PAM must be selected.
When it comes to CRISPR experiments, therefore, you often must make a choice about what is your top priority: location or efficiency. If you are just trying to shred a long non-coding RNA, cutting most anywhere within the RNA will do. Similarly, if you want to knock out a gene, a frameshift mutation introduced anywhere within the early coding region will do the job. What matters for these experiments is cutting efficiency so that the greatest effect possible will be achieved. On the other hand, if you are trying to knock-in a patient mutation or need to make a precise amino acid change, there is very little flexibility in the location of the cut and subsequent edit. In these applications, location takes priority over cutting efficiency — although both are of course preferred!
When trying to strike that balance, you may wonder: how close is close enough for the desired edit to still occur if the cut site is sub-optimal? As with everything in biology, the answer is always it depends, but generally the efficiency of the desired edit is reduced by over 50% when the cut site is just 10 bps away from the site in question (Kwart et al, 2017). To add insult to injury, the most utilized PAM sequence, that of SpCas9, is NGG. Assuming all nucleotides are represented equally, that’s only a 1/16 chance of having a PAM at a given site. While it is possible to "get lucky" and find a great gRNA with a proximal PAM right where you need it, the odds of this occurring are not in your favor. Thus, there is a need to eliminate or reduce the PAM requirement to allow flexibility in gRNA selection and subsequent editing.
Identifying PAM-free and -flexible Cas enzymes
The solution to the PAM problem seems simple, in theory: find a Cas enzyme or evolve one that uses a PAM with no sequence constraints, or at least has looser or different requirements. Luckily, there are some available.
Cas proteins that don’t utilize NGG
There are plenty of Cas9 variants which utilize alternative PAMs. These alternative PAMs are not necessarily more flexible than NGG; they’re often just different (e.g. NGAG, NGAN, etc.). Additionally, alternative Cas enzymes exist, such as Cpf1/Cas12a, which recognizes the PAM sequences of TTTV. While all of these enzymes certainly add more PAM options, none of them offer a one size fits all solution to the issue. Nor are all Cas enzymes equal; just because an enzyme recognizes a PAM sequence in your site doesn’t mean it’s suitable for your experiment.
If you’re looking for more pan-genome PAM flexibility, you’ll want to consider one of several Cas9s that recognize NG PAMs. The Nureki lab generated SpCas9-NG, a rationally engineered Cas9 with NG PAM compatibility, as the name implies (Nishimasu et. al, 2018). xCas9 was generated by the Liu lab through phage-assisted evolution, specifically with the goal of generating Cas enzymes with alternative PAM requirements (Zhao et. al, 2023). xCas9 has been shown to be less effective at NGH PAMs, though (Zhang et al, & Walton et al). Additionally, SpG Cas9 with the same PAM preference of NG was isolated by the Kleinstiver Lab (read more on this lab's other developments below!) (Walton et. al, 2020). These Cas proteins have everything we know and love about Cas9, but with broader PAM compatibility. This alternative PAM requirement adds a significant amount of flexibility compared to a strict single-compatibility sequence like NGG. Nonetheless, in an AT-rich region, these Cas9s may still leave you choosing between efficiency and location.
Nearly PAM-less editing
The Kleinstiver lab sought to address the PAM problem through structure-guided engineering of the PAM-interacting domain of Cas9. After multiple rounds of engineering, they developed the SpRY Cas9 variant (Walton, et. al, 2020). SpRY uses NRN PAMs and is capable of editing at frequencies comparable to SpCas9. SpRY can also edit NYN PAMs, but with less efficiency and less reliability.
Notably, the SpRY variant didn’t display higher off-target effects than SpCas9, a pleasant surprise since the PAM flexibility could in theory promote off-target binding. The activity of SpRY was validated in double stranded cleavage and base editing applications, with high editing capacity in both contexts for NRN PAMs and to a lesser extent NYN. This increases the flexibility of gRNA placement significantly over SpCas9 and more so than Cas9s with an NG PAM.
| Cas | PAM sequence | Additional notes |
| NGG | - | |
| TTTV | Great for AT-rich regions | |
| NGAG | - | |
| NG |
Less effective at NGH PAMs (Zhang et al, 2021) |
|
| NG | - | |
| NG | - | |
|
|
NYN/NRN | NYN is less effective than NRN |
Table 1 - Summary of PAM-flexible Cas enzymes
Are we truly PAM-less?
Have we reached an essentially PAM-free solution? Almost! Assuming a PAM of NRN, 50% of randomly selected genomic loci will have a PAM located next to it. The SpRY Cas9 can also work with NYN PAMs; it just isn’t always ideal. But the increased flexibility helps even when it’s not an ideal site: moving several bps away from a desired site does not have the same devastating impact on efficiency as 10-15 bps away does. This Cas thus allows precise targeting (give or take a bp or two upstream or downstream) for most gene targeting experiments. Between SpRY and several of the other Cas variants described, nearly all genomic landscapes should be editable with CRISPR technology.
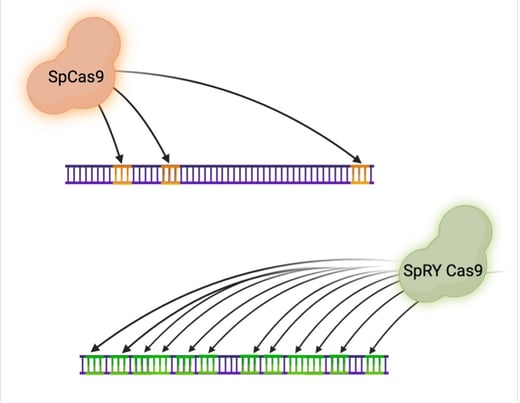
|
| Fig. 1: SpCas9 has sparse PAM coverage in the human genome. SpRY Cas9 has PAM sites covering the majority of the genome! |
Want to get your hands on SpRY? Find a mammalian plasmid here and edit on!
References and Resources
References
Kwart, D., Paquet, D., Teo, S. et al. Precise and efficient scarless genome editing in stem cells using CORRECT. Nat Protoc 12, 329–354 (2017). DOI:10.1038/nprot.2016.171
Zhao, L., Koseki, S.R.T., Silverstein, R.A. et al. PAM-flexible genome editing with an engineered chimeric Cas9. Nat Commun 14, 6175 (2023). DOI:10.1038/s41467-023-41829-y (Plasmids available here)
Walton, R. T., et al., Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368,290-296(2020). DOI:10.1126/science.aba8853 (Plasmids available here)
Resources on Addgene.org
- Addgene’s CRISPR Guide
- CRISPR Plasmids and Resources
Resources on the Addgene blog
- The PAM Requirement and Expanding Beyond Cas9
- xCas9: Engineering a CRISPR Variant with PAM Flexibility
- History of CRISPR Cas – A Tale of Survival and Evolution
Topics: CRISPR, CRISPR 101






Leave a Comment