Turn it on? Turn it off? Turn it down, but not forever? You don’t need to resort to the force to control protein expression in mammalian cells: easy to use biological tools are abundant! In this post, we will review tried-and-true protein expression control tools as well as explore some new innovations. Whether you want to turn things on or off, permanently or temporarily, we’ve got you covered.
Defining your control requirements
To pick the best tool for the job, it’s important to know what the job requirements are. Here are the questions you should be asking.
What kind of control? On, off, tunable, and/or inducible?
How long do you need control? Will temporary control (a few days) be enough, or do you want a permanent control mechanism?
Where is the control happening? Is the regulation of an exogenously introduced gene or over the native locus?
Let’s explore each of the options together.
Turn it off
Turn it off forever
If you want to turn off protein expression, you are almost certainly working with an endogenous locus. The most straightforward way to turn off expression permanently is to knock out the gene for your protein of interest. CRISPR-Cas9 and a custom gRNA is all you need to generate a frameshift mutation that will permanently ablate protein expression.
If you need the ability to control when you turn off expression for good, the Cre-Lox system offers inducible yet permanent gene excision. Essentially, LoxP sites are knocked in flanking a gene of interest (or a single exon) and everything in between those sites can be excised by intentional introduction of the Cre recombinase. This is a time-tested system but does require knock-in of the LoxP sites, which is more difficult than a simple knock out.
Looking for some permanent knock out plasmids? Check out our Cre vector and Murine LoxP vector.
Turn it off for a little while
Transient suppression of expression is faster and easier than making a knockout and is a great way to temporarily reduce expression of essential proteins (when knockouts aren’t viable). siRNA or RNAi are small, double stranded RNA molecules that degrade target mRNA transcripts. When transiently introduced, they can deplete endogenous protein levels for 1-3 days.
There’s also a CRISPR for that! CRISPR interference (CRISPRi) targets a catalytically dead Cas9 fused to a transcriptional repressor to your gene of interest. It will inhibit transcription instead of introducing a DNA double strand break. Transient introduction of CRISPRi or inducible expression (more on this below) will both achieve temporary repression of protein expression.
Addgene has a dCas9 KRAB plasmid for transient knockouts!
Turn it on
Turn it on forever
Permanently increasing or turning on expression of a new protein is a cinch. Stably introducing the coding region of a gene (cDNA) can be done by either plasmid or virus. A plasmid carrying your cDNA can be targeted for integration at a specific genomic site, or randomly integrated (once or many times) into the genome. Make sure to choose an appropriate promoter to express your cDNA, depending on the level of expression you would like. Check out our promoter blog if you need help choosing.
Viral expression is another mechanism to integrate your choice cDNA. Both lentivirus and retrovirus can package cDNAs for genomic delivery and have many therapeutic advantages, such as tissue-specific targeting.
Looking to permanently induce expression? Check out our Retro expression vector and Lenti expression vector.
Turn it on for just a little while
Yes, there’s a CRISPR for that, too! CRISPR activators (CRISPRa) are transcriptional activators fused to a dead Cas9. The system is led, via a custom gRNA, to your gene of interest to temporarily increase expression. There are several flavors of activators which can be used with this approach - read more about them on our CRISPRa blog!
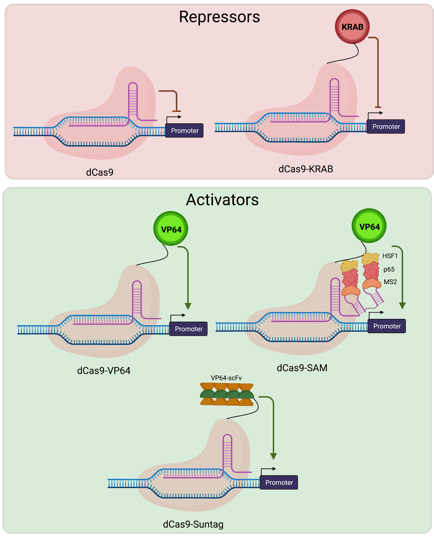 |
|
Fig. 1 CRISPR activator and repressor mechanisms |
Want to transiently induce expression? Check out Addgene's Lentiviral dCas9 SAM vector and Piggybac dCas9 SAM vector!
On/off control all the time
If your experimental system requires tunable control - or maybe you’re just a control freak - this section is for you! Many of the tools highlighted in this section can be used to turn expression on/off for short or long periods of time. Thus, there is a lot of overlap with the systems discussed above, just with more elements of control.
Tet promoters
Tetracycline-dependent promoters are engineered by placing tet response elements upstream of a minimal promoter. These systems come in two varieties: tet-on and tet-off. As the names imply, tet-off turns off constitutively expressed proteins while tet-on turns on constitutively silenced proteins. Addition of tetracycline or one of its analogs will induce these transcriptional changes, and expression is tunable by titration with different amounts of tetracycline. Tetracycline regulation is reversible (when the drug is removed expression goes back to baseline) and occurs at the transcriptional level.
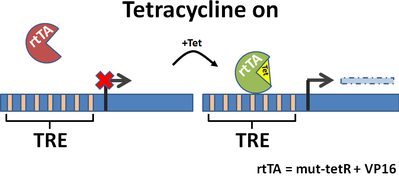
|
Fig. 2 Tetracycline on/off promoters and modes of action
Check out Addgene's Lentiviral Tet-on and AAVS1 targeted Tet-on transgene vector.
Degron tags
Tagging relevant proteins with a degron tag can provide inducible protein degradation. These systems work by tagging the endogenously or exogenously expressed protein with the relevant tag and then introducing a specific ligand to mark the tag and whatever it is fused to for degradation. Since the tagged protein of interest is normally expressed, protein levels will quickly recover once the ligand is removed. Degron tag systems include small molecule assisted shut off (SMASh), auxin-inducible degron (AID), and degradation tag (dTAG). These control systems function at the post-translational level, unlike most of the other mechanisms described.
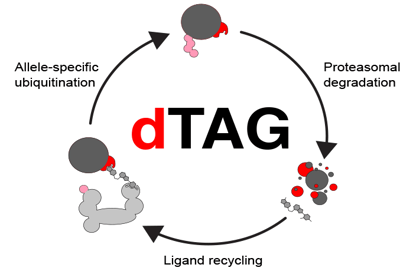 |
|
Fig. 3 dTag mechanism of protein degradation. Adapted from Nabet et al. |
Check out Addgene's Lentiviral dTAG cloning vector and SMASh degron tagging vector!
Destabilizing domains
Destabilizing domains (DD) are akin to degron tags, just with a flipped mechanism. When a protein is tagged with a DD, it is unstable and quickly degraded. However, if the relevant small molecule is introduced into the system, it will bind and stabilize the DD, resulting in sustained protein levels.
Looking for a DD? Check out our DD-Cas9 vector!
The price of tunable control
These systems can be easily implemented for exogenously introduced proteins, as many of them just need a special promoter or tag prior to introduction of the construct into cells. However, if you would like tunable control of an endogenously expressed protein, then these systems need to be site-specifically knocked in. This can be labor intensive and time consuming. If your native protein doesn’t require this level of control, one of the previous options may be better suited for the job.
Whether you want to express or repress, good luck on your protein expression journey!
Resources and References
References
Nabet et al. The dTAG system for immediate and target-specific protein degradation. Nature Chemical Biology. May 2018. 14(5): 431-441. PubMed PMID: 29581585.
Additional resources on the Addgene blog
- Plasmid 101: Inducible Promoters
- Plasmid 101: Repressible Promoters
- CRISPR 101: Which Cas9 Do I Choose for my Experiment
Additional resources on the Addgene website
Topics: Plasmids





Leave a Comment