Taking the road less traveled and generating a knock-in cell line instead of a knock-out? We’ve got you covered in this blog, with tips and tricks for harnessing the homology-directed repair pathway, designing the best donor DNA, and avoiding common mishaps in this class of genome edit.
Knock-in mutations and gene editing
Targeted genome editing events fall into two general classes: knock-in and knock-out. The goal of a knock-out is to disrupt DNA translation by generating a frameshift mutation. The majority of cellular repair events will generate these types of mutations, making it an ‘easy’ editing job to get done. However, this is not the case for knock-ins. Knock-in mutations typically require an exact DNA sequence to be incorporated at a precise genomic location, with little to no room for error. It’s no surprise then that these edits take more planning and finesse then their knock-out counterparts.
To kick off a knock-in experiment the first questions are where, which, and what. Identify where in the genome you would like to introduce your knock-in. Then choose which Cas enzyme to use and design a gRNA to where you would like to introduce the edit. Finally, you must stipulate what sequence to introduce, which is done through the design and use of a donor DNA molecule. Your donor molecule will need to have homology to the targeted locus on either side of the new sequence. This homology will guide the cell to use the donor DNA as a template for repair, which will result in the incorporation of your sequence, a.k.a a knock-in! So how does the donor get used as a template for repair? Read on to find out!
Homology-directed repair and genome engineering
The easiest way to make a knock-in cell line is to utilize the built-in repair pathway cells already have for repairing DNA double strand breaks – (HR).
HR is the dominant homology-directed repair (HDR) pathway in mammalian cells, but a less frequent DNA repair mechanism in general. On average, it accounts for less than 10% of DNA repair, with this rate varying across cell types. Variance can be caused by cell cycle rates; rapidly diving cells will have higher HR frequencies than quiescent cells. It is important to know that HR is often mutated in cancer cells, specifically in breast and ovarian lines commonly used in lab. It is recommended you check your cell lines for HR-related mutations before attempting knock-ins by HDR.
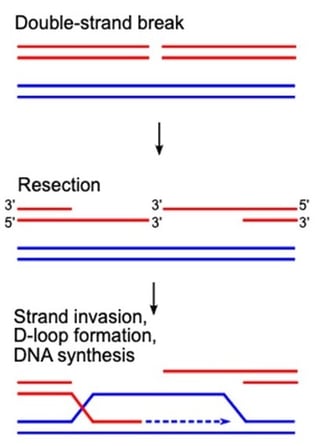
|
| Fig. 1: Early steps in repair of a DNA double strand break by homologous recombination. |
Designing donors for HDR
The basis of the HDR pathway relies on repair from a template molecule, which is usually an endogenously available sister chromatid, though exogenous DNA can be used as well. Below are some considerations for designing donor DNA for successful HDR events.
CRISPR cut site location
Position your CRISPR cut site as close as possible to the knock-in location. HDR depends on resection at the break site to form ssDNA and uses the ends of the ssDNA to find a repair template. The highest HDR efficiency is observed when inserts are within 10 bps of the break, making the cut off essentially as part of the templated DNA. Check out our blog post on increasing HDR efficiency to read more about optimizing this distance.
Homology arms and types of donors
To make sure your donor molecule is used as the template, you need to incorporate homologous sequence to the right and/or left of the knock-in site. The length of that sequence and the makeup (single vs. double stranded) all depends on the size of the knock-in. Single strand oligonucleotide donors (ssODNs) with homology arms as short as 40 bps can efficiently knock-in a few hundred bps of sequence. However, if you have a larger knock-in (200 bp – 2 kb), a dsDNA donor will need to be used due to synthesis limits of oligonucleotides. These donors traditionally have larger homology arms in the 500 bp to 1 kb range, however, shorter homology tracts have recently been used with some success (Yu et al., Nat Chem Biol).
Mutate the PAM or sgRNA sequences
Cas9 likely goes through multiple rounds of cleavage at the target site until either the PAM site or part of the guide’s recognition sequence is destroyed via mutation. By introducing a mutation in either the PAM or the guide recognition sequence in the donor DNA, you can guarantee there won’t be re-targeting after HDR. If your Cas9 cuts in the coding region of a gene, make sure that the PAM edit you introduce is a silent mutation, so you don’t accidentally change your protein of interest.
-min.png?width=770&height=454&name=image%20(2)-min.png)
|
| Fig. 2: Stop re-cutting with PAM and guide RNA disrupting mutations in donor DNA |
Promoting HDR events over other edits
Like I said earlier, HDR is not the most common editing pathway for DNA repair. To tilt the scales in your favor for an HDR edit, you can try one or more of the below suggestions.
Disable the other repair mechanisms
Mammalian cells have 2 primary repair mechanisms besides HR - Non-homologous end-joining (NHEJ) and alternative end joining (alt-EJ). Inhibitors to essential components of both pathways have been shown to be efficacious for increasing knock-in events (Yang et al., Int J Mol Sci). Adding these inhibitors to your culture media prior to and after Cas9 introduction increases HDR, just make sure to remove the inhibitors within a day or two to not affect the viability of your cells.
Enrich S-phase cells
HR operates predominantly in S and G2 phase of the cell cycle, when a sister chromatid is likely to exist as a repair template. To maximize HDR events, ensure cells are actively cycling through the cell cycle stages. If cells are overly confluent or don’t have sufficient nutrients in their media, they will not undergo DNA replication, so make sure you are keeping an eye on those cells! To take this one step further, you can inhibit cell cycle regulatory proteins responsible for the transition out of S phase, so cells stay in an HDR-promoted cycle longer. This approach may have some negative outcomes on viability if cells are arrested too long. Prolonged cell cycle arrest can lead to apoptosis in as little as 10 hours, so read up on the timing of this approach carefully (Lin et al., eLife)!
Use a Cas that facilitates HR
What if promoting HR was as simple as swapping out your Cas9? It can be! Cas12a is a Cas family member which generates sticky ends upon cutting. Cas12a may promote HDR by generating ssDNA overhangs at the break site – an early HR step (Moreno-Mateos, et al., Nat Com).
Another method used to prime a Cas break for HR is to fuse an HR protein to the nuclease. Early HR factors fused to Cas9 have been shown to increase HDR frequency by committing a break to performing HR before other factors were loaded onto the break, potentially shuttling repair down a different pathway (Charpentier et al., Nat Com).
-min.png?width=697&height=428&name=image%20(1)-min.png)
|
| Fig. 3: HDR events compete with NHEJ-mediated edits of small insertions and deletions. |
Start your knock-in experiment today!
Get ready to identify knock-in clones by reading up on validating genome edits to find screening strategies for your experiment. Also learn about Addgene’s knock-in plasmids for reagents to help you knock your knock-in experiment out of the park. Happy editing!
Resources and References
References
Yu, Y., Guo, Y., Tian, Qigi, Lan, Y., et. al. An efficient gene knock-in strategy using 5’-modified dsDNA donors with short homology arms. Nat Chem Biol., 308(20): 1-9 (2020). 10.1038/s41589-019-0432-1
Mehdi Banan. Recent advances in CRISPR/Cas9-mediated knock-ins in mammalian cells. J Biotechnol., 16(4): 387-390 (2020). 10.1016/j.jbiotec.2019.11.010
Wright, D. W., Shah, S. S., Heyer, W. D. Homologous recombination and the repair of DNA double strand breaks. J Biol Chem., 293(27): 10524-10535 (2018). 1 10.1074/jbc.TM118.000372
Yang, H., Ren, S., Yu, S., Pan, H., et al. Methods favoring homology-directed repair choice in response to CRISPR/Cas9 Induced-double strand breaks. Int J Mol Sci., 21(18): 6461 (2020). 10.3390/ijms21186461
Lin, S., Staahl, B. T., Alla, R. K., Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLIFE. (2014) 10.7554/eLife.04766
Moreno-Mateos, M. A., Fernandez, J. P., Rouet, R., Vejnar, C. E., et al. CRISPR-Cpf1 mediates efficient homology-directed repair and termperature-controlled genome editing. Nat. Com., 8(2024), (2017). 10.1038/s41467-017-01836-2
Charpentier, M., Khedher, A. H. Y., Menerot, S., Brion, A., et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Com., 9-1133 (2018). 10.3390/ijms21186461
Additional Resources on the Addgene Blog
- Check out our CRISPR Featured Topic Page
- Learn about Non-Homologous End Joining
- Learn about Base Editing
- Read 3 Tips to Improve HDR Efficiency for CRISPR Editing in Human Cells
Resources on Addgene.org
- Browse All CRISPR Plasmids
- Find Validated gRNAs for Your Next Experiment
- Check out our CRISPR Guide
Topics: CRISPR 101






Leave a Comment