Rosella is a pH-sensitive fluorescent biosensor that was recently deposited with Addgene by Dr. Mark Prescott. This system was developed for monitoring and analyzing autophagy of cytosol and organelles in yeast cells. Autophagy (Greek for “self-eating”) is induced by a lack of nutrients and targets cytosol and organelles to the vacuole/lysosome for degradation and recycling. The key to Rosella’s autophagy-sensing abilities is that its fluorescence emission spectra changes when it goes from a more neutral pH compartment, like the cytosol, to the higher pH of the vacuole. Read on to learn more about prior methods for studying autophagy and how Rosella improves upon them.

Methods for studying autophagy
Biochemical assays
-
Long-lived protein degradation: This approach measures the degradation rate of radio-labeled long-lived proteins as a proxy for autophagy. Cells are cultured with isotope-labeled amino acids for several hours to several days, followed by a short period of growth with unlabeled amino acids. This washout period removes radio-labeled short-lived proteins primarily via proteasome degradation. Autophagy is then induced and quantified by measuring the amount of radioactivity in the culture supernatant (this is indicative of protein degradation). To control for protein degradation due to other pathways, it is standard practice to compare degradation rates between samples treated with or without an autophagy inhibitor. While this method does provide a quantitative measure of autophagy, it only measures bulk autophagy (as opposed to giving more fine-grained detail about what proteins or organelles are being degraded) and it is slow.
-
Alkaline phosphatase activity: In yeast, the PHO8 gene encodes a vacuolar alkaline phosphatase. Normally PHO8 is synthesized at the ER, delivered to the vacuole via the secretory pathway, and then cleaved to generate an active form of the protein. To monitor autophagy, the Pho8Δ60 mutant protein is expressed. Pho8Δ60 localizes to the cytosol in an inactive form unless autophagy is induced. Then non-selective macroautophagy of the cytosol leads to accumulation of Pho8Δ60 in the vacuole, where it is cleaved to generate an active enzyme. Pho8 phosphatase activity or the molecular weight shift of Pho8 from its uncleaved to cleaved form is the final read out of this assay. This is also a quantitative measure of autophagy, but, like the protein degradation assay above, it’s slow and only measures bulk autophagy.
Morphological assays
-
Electron microscopy: Electron microscopy is a traditional method for studying autophagy. This method relies on the identification of autophagic structures based on morphology. Autophagosomes are relatively easy to identify: double-membraned structures containing undigested cytoplasmic contents. Autophagosomes that have fused with the vacuole or a lysosome are trickier to identify because their contents can be at various stages of degradation. Additionally, electron microscopy analysis is time consuming.
-
GFP-tagged Atg8p or LC3: ATG8 is part of a group of genes that affect autophagy (ATG) in yeast. Atg8p (“p” stands for protein) associates with autophagosomal membranes, so tagging it with GFP allows for tracking the localization or accumulation of pre-autophagosomal structures, autophagosomes and autophagic bodies in yeast. A GFP-tagged version of LC3, the mammalian Atg8p homolog, can also be used to monitor autophagy, but under some conditions it aggregates in an autophagy-independent manner. Additionally, tracking Atg8p or LC3 doesn’t provide information about the contents of the autophagosome nor does it provide information about what is being degraded in the vacuole or lysosome, since both Atg8p and LC3 are degraded or released once an autophagosome fuses with the vacuole or lysosome.
Rosella fluorescent properties
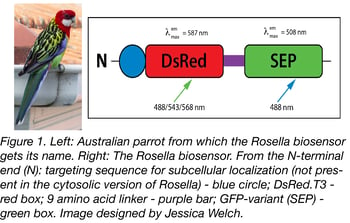
Rosella is a dual color-emission biosensor named after the brightly-coloured Australian parrot. It’s comprised of two tandem fluorescent proteins: a relatively pH-insensitive RFP variant, DsRed.T3 and a pH-sensitive GFP variant, super ecliptic pHluorin (SEP). They are connected by a 9 amino acid linker that’s fused to the C-terminus of DsRed.T3. See Table 1 for a summary of Rosella’s excitation and emission spectra. DsRed is the permanent fluorescent tag portion of Rosella: it will emit red fluorescence regardless of its localization in the cell. SEP’s pH sensitivity means Rosella will fluoresce green unless it’s in an acidic environment like the vacuole or lysosome. See Table 2 for a summary of what color fluorescence Rosella emits based on its localization.
Table 1: Summary of Rosella’s excitation and emission spectra.
| Component | Excitation | Emission | pH Range |
| DsRed.T3 |
488, 543*, 568 nm |
587 nm |
~4.9 - 9 |
| SEP |
488* nm |
508 nm |
~6.5 - 9 |
*Optimal sequential excitations wavelengths as determined by Rosado et al: see Table 2 for more details on the effect of pH on the fluorescent properties of Rosella.
Table 2: Effect of Localization on Rosella’s fluorescence emission.
| Localization | pH | Emission Color |
| Cytosol |
~7.5 |
Red and Green |
| Mitochondria |
~8 |
Red and Green |
| Vacuole |
~6.2 |
Red |
| Lysosome | ~4.8 | Red |
Using Rosella to study autophagy in yeast
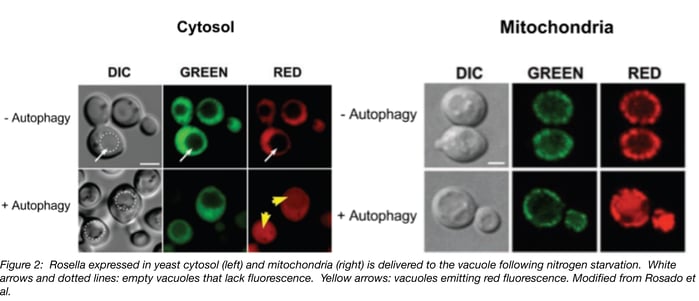
Rosella is targeted to the cytosol when expressed without a signal sequence and to the mitochondria via citrate synthase’s mitochondrial targeting sequence. Under normal growth conditions, both variants are excluded from the vacuole and emit red and green fluorescence. After 4 hours of nitrogen starvation to induce autophagy, red but not green fluorescence accumulates in the vacuole. See Figure 2 for an example of what fluorescence looks like for cytoplasmic mitochondrial Rosella with and without autophagy. In both cases, red fluorescence in the vacuole increased with longer exposure to autophagy-inducing conditions.
Rosella makes it easier to study the mechanisms behind autophagy by tracking what’s being transported to the yeast vacuole (i.e. cytosol, mitochondria), rather than tracking generic autophagy markers. It’s particularly useful for comparing bulk autophagy vs. mitophagy, the targeted autophagy of mitochondria. As shown in Sargsyan et al. It is also possible to make Rosella fusions and thereby monitor autophagy of your protein of interest.
Are you ready to try out Rosella? Both the cytosol-targeted and mitochondria-targeted variants are available from Addgene.
References
1. Rosado, C., Mijaljica, D., Hatzinisiriou, I., Prescott, M., & Devenish, R. J. (2008). Rosella: A fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy,4(2), 205-213. doi:10.4161/auto.5331. PubMed PMID: 18094608
2. Sargsyan, A., Cai, J., Fandino, L. B., Labasky, M. E., Forostyan, T., Colosimo, L. K., Graham, T. E. (2015). Rapid parallel measurements of macroautophagy and mitophagy in mammalian cells using a single fluorescent biosensor. Scientific Reports,5(1). doi:10.1038/srep12397. PubMed PMID: 26215030. PubMed Central PMCID: PMC4517063.
3. Mizushima, N., Yoshimori, T., & Levine, B. (2010). Methods in Mammalian Autophagy Research. Cell,140(3), 313-326. doi:10.1016/j.cell.2010.01.028. PubMed PMID: 20144757. PubMed Central PMCID: PMC2852113.
4. Klionsky, D. J., Cuervo, A. M., & Seglen, P. O. (2007). Methods for Monitoring Autophagy from Yeast to Human. Autophagy,3(3), 181-206. doi:10.4161/auto.3678. PubMed PMID:17224625.
5. Noda, T., & Klionsky, D. J. (2008). Chapter 3 The Quantitative Pho8Δ60 Assay of Nonspecific Autophagy. Methods in Enzymology Autophagy: Lower Eukaryotes and Non-Mammalian Systems, Part A, 33-42. doi:10.1016/s0076-6879(08)03203-5. PubMed PMID: 19185711.
Additional Resources on the Addgene Blog
- Beware of Fluorescent Protein Oligomerization
- Seeing Red: Simple GFP Photoconversion
- Light Sheet Fluorescence Microscopy
Additional Resources on Addgene.org
- Fluorescent Protein Plasmids and Resources
- Fluorescent Protein Guide: Biosensors
- Keima, another fluorescent pH-biosensor for studying autophagy





Leave a Comment