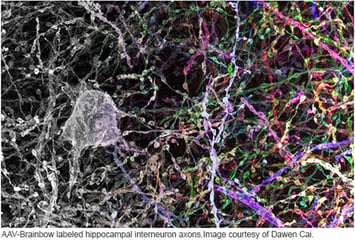
 CRISPR-Cas9 genome editing may be the hot new way to manipulate gene expression, but other gene manipulation systems remain valuable to biology. Cre-lox recombination, discovered in the 1980s, is one of the most important ways to spatially and temporally control gene expression, especially in in vivo models, and new Cre-lox based technologies are still being developed today. In this post, I will highlight the evolution of the Brainbow multicolor labeling system - a perfect example of the continued utility of Cre-lox. Check out our previous blog post, Plasmids 101: Cre-lox, if you need a quick primer on how Cre-lox recombination works.
CRISPR-Cas9 genome editing may be the hot new way to manipulate gene expression, but other gene manipulation systems remain valuable to biology. Cre-lox recombination, discovered in the 1980s, is one of the most important ways to spatially and temporally control gene expression, especially in in vivo models, and new Cre-lox based technologies are still being developed today. In this post, I will highlight the evolution of the Brainbow multicolor labeling system - a perfect example of the continued utility of Cre-lox. Check out our previous blog post, Plasmids 101: Cre-lox, if you need a quick primer on how Cre-lox recombination works.
Advances in Cre-lox recombination have included inducible Cre, such as the tamoxifen-responsive CreERT, the coupling of Cre-lox with FLP-FRT recombination to generate additional recombination events, and the synthesis of alternative loxP sites to specify distinct patterns of recombination. In 2007, Joshua Sanes and Jeff Lichtman, both of Harvard’s Department of Molecular and Cellular Biology, harnessed these attributes of Cre-lox to create the Brainbow mouse neural labeling system. Brainbow-1 and -2 have been further refined to create Brainbow 3.2. Brainbow labeling has also been applied to cell marking and lineage tracing in other systems, such as zebrafish and Drosophila.
Why Brainbow?
Brainbow highlights the trend in biology towards single-cell resolution imaging. Sanes and Lichtman endeavored to create a map of the brain, a connectome that would detail how neurons form circuits and where they synapse. View the overview schematic at the bottom of this post for a basic synopsis of the differences amongst the Brainbow systems. Pre-Brainbow techniques, such as Golgi staining, injection of diffusible labels, or electron microscopy of stained sections, were limited due to low resolution or were very time-intensive, necessitating the development of a new technique. In a structure as tightly packed and interwoven as the brain, robust single-cell labeling is necessary to distinguish individual cells.
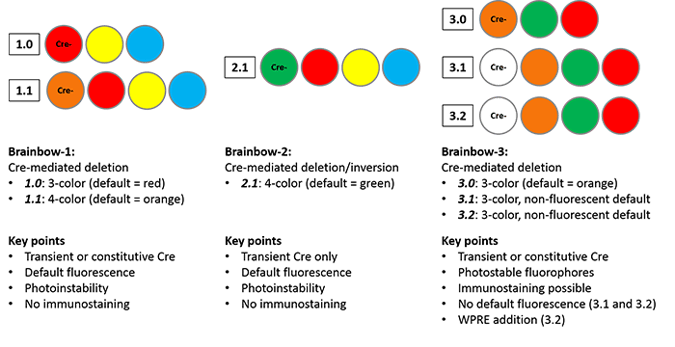
The Brainbow-1.0 construct contains three fluorescent proteins: RFP (red), YFP (yellow) and M-CFP (membrane tethered cyan). Without Cre recombination, RFP is expressed. Cre can mediate one of two deletions to allow YFP or M-CFP expression; these deletions are defined using 2 loxP variants (only identical loxP sites can mediate recombination.) The deletions are mutually exclusive, as either deletion removes one of the other loxP site variants, preventing further recombination. Brainbow-1.1 scales up the system to four fluorescent proteins, with recombination events specified by three loxP variants, but the principles remain the same. The first fluorescent protein in the construct, OFP (orange), is expressed when Cre is absent; stochastic Cre excision results in one of three recombinations, and subsequent recombinations cannot occur.
While the Brainbow-1 system employs Cre-mediated deletion, the design of Brainbow-2.1 cleverly combines Cre-mediated deletion and inversion. The Brainbow-2.1 construct can express one of four colors (n-GFP, RFP, YFP or M-CFP.) The construct contains two tandem invertible segments, each with two fluorescent protein coding sequences in opposite orientations. Cre-mediated excision removes one segment, eliminating two color possibilities. The remaining cassette can invert as long as Cre remains expressed; once Cre is removed from the system, the fluorescent protein whose coding sequence is closest to the promoter will be expressed. This system differs from Brainbow-1.0 in that transient Cre recombination is required to prevent constant inversion of the construct.
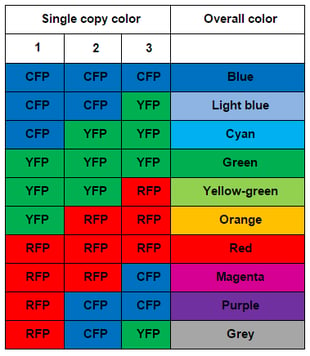
Sanes and Lichtman used pronuclear injection to insert these Brainbow constructs into mice. When they imaged the mouse brains, they found many more than 3-4 colors due to the tandem integration of multiple copies of the construct (about 8 in Brainbow-1.0 mice.) The combinatorial effect of multiple, independent recombination events leads to a rainbow of colors. With three copies of the construct, one would expect ten colors (see table to the right); in various reports, Sanes and Lichtman have observed from 90-160 distinct colors due to higher copy number. The name Brainbow is truly fitting for this colorful technology.
Optimizing the system: Brainbow-3
Although Brainbow provided neuroscientists with the vast array of colors needed to mark individual neurons, the system also suffered from a number of limitations. First, the imaging of Brainbow mouse tissue was challenging due to low fluorescence intensity, caused partially by fluorophore photoinstability, as well as the tendency of fluorophores to aggregate in the neurons’ somata. Second, the system did not permit analysis by immunostaining. Although the fluorophores used are fluorescently distinct, their protein sequences are highly homologous, preventing design of antibodies specific to each fluorophore. Third, Brainbow-1 and Brainbow-2 each contained a “default” state; for example, Brainbow-1.0 expresses RFP when the construct has not undergone recombination. This default state was disproportionately expressed by a majority of neurons, limiting the number of distinct colors that could be observed in a given area.
In 2013, Dawen Cai, et al. released a refinement of the Brainbow technology, Brainbow-3.0, with the goal of overcoming the limitations listed above. First, they screened a variety of fluorescent proteins to find those with ideal characteristics (low aggregation, high photostability, and high stability after fixation.) From the seven resulting proteins, they chose three with low levels of both fluorescence and sequence overlap (coral mOrange2, jellyfish EGFP, and sea anemone mKate2.) They then successfully generated custom antibodies to each of these proteins and confirmed that there was no cross-reactivity, opening the door for immunostaining analysis. To achieve even cell labeling, they generated farnesylated derivatives of the fluorescent proteins, which are directly trafficked to cell membranes. Membrane trafficking of farnesylated derivatives enables labeling of delicate axonal and dendritic processes not previously visible with Brainbow-1 and -2.
The general structure of Brainbow-1.0 is retained in Brainbow-3.0, but with mOrange2, EGFP and mKate2 as the fluorophores; mOrange2 is the default state. In contrast, Brainbow-3.1 and -3.2 do not display default fluorescence due to the addition of translation-blocking STOP cassette immediately following the promoter. The STOP cassette includes a mutant YFP that does not fluoresce, but can be detected via immunostaining. This feature facilitates screening of Cre-negative Brainbow mice to determine the number and type of cells in which the construct is expressed. The photostability of Brainbow-3.2 is improved due to the addition of a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), commonly used to increase transgene protein levels.
Overview of Variations Between Brainbow Systems
Brainbow variants and applications beyond the mouse brain
In addition to improving the Brainbow system, Sanes and Lichtman also developed a complementary Flpbow construct that is functionally similar to Cre-based Brainbow, but is controlled by FLP/FRT recombinase. When placed under different promoters, Brainbow and Flpbow can be used to label distinct cell populations.
To decrease the animal breeding necessary to produce animals with Brainbow transgenes and Cre, Cai et al. also created an Autobow construct containing both Cre and XFPs. Cre production drives recombination and XFP selection, followed by Cre self-excision. These constructs are stably maintained through at least six generations.
In addition to the neuronal pThy1-Brainbow constructs, Addgene also has two Brainbow adeno-associated viral vectors (AAV) available - AAV-EF1a-BbChT and AAV-EF1a-BbTagBY. These constructs contain two XFPs each due to size limitations associated with AAV; co-infection with both constructs produces a minimum of 8 colors. The use of AAV provides spatial and temporal control without the need for germline modification, and enables Brainbow to be used in a variety of species.
More variants have been created by other labs, including R26R-Confetti described in Hugo J. Snippert, et al. (2010) and the MAGIC Marker strategy described in Karine Loulier, et al. (2014).
Currently, Brainbow techniques are also being applied to study model organisms such as Drosophila and zebrafish. Brainbow in Drosophila has aided in the mapping of neural circuits, such as connections between motor neurons and the neuromuscular junction. In zebrafish, the method has become very useful in lineage tracing; “Zebrabow” was used to trace the development of the corneal epithelium.
For scientists interested in neuroscience and development, Brainbow is a valuable tool to mark and follow single cells due to the wide array and high stability of colors. Sanes and Lichtman estimate that Brainbow’s colorful labeling has decreased the mapping time for a given section of brain by at least an order of magnitude. It is clear that further refinements of the Brainbow technique will provide important insights into the complicated physical organization of the brain and other biological systems.
Interested in applying Brainbow to your research? Check out Brainbow plasmids available from Addgene!
Find Brainbow Plasmids @Addgene:
References:
Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Livet J, Weissman TA, Kang H, Draft RW, Bennis RA, Sanes JR, Lichtman JW. Nature. 2007 Nov 1;450(7166):56-62. PubMed.
A technicolour approach to the connectome. Lichtman JW, Livet J, Sanes JR. Nature Reviews Neuroscience. 2008 Jun;9(6):417-22. doi: 10.1038/nrn2391. Epub 2008 Apr 30. PubMed.
Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Cell. 2010 Oct 1;143(1):134-44. PubMed.
Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Hampel S, Chung P, McKellar CE, Hall D, Looger LL, Simpson JH. Nature Methods. 2011 Mar;8(3):253-9. doi: 10.1038/nmeth.1566. Epub 2011 Feb 6. PubMed.
Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, Salecker I. Nature Methods. 2011 Mar;8(3):260-6. doi: 10.1038/nmeth.1567. Epub 2011 Feb 6. PubMed.
Improved tools for the Brainbow toolbox. Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Nature Methods. 2013 May 5:10(6):540-7. doi: 10.1038/nmeth.2450. Epub 2013 May 5. PubMed.
Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, Ciruna B, Sanes JR, Luchtman JW, Schier AF. Development. 2013 Jul;140(13):2835-46. doi: 10.1242/dev.094631. PubMed.
Multiplex cell and lineage tracking with combinatorial labels. Loulier K, Barry R, Mahou P, Le Franc Y, Supatto W, Matho KS, Ieng S, Fouquet S, Dupin E, Benosman R, Chédotal A, Beaurepaire E, Morin X, Livet J. Neuron. 2014 Feb 5;81(3):505-20. PubMed.
Topics: Fluorescent Proteins, Fluorescent Imaging, Cell Tracing, Neuroscience







Leave a Comment