THE ORIGINAL NgAgo ARTICLE DISCUSSED IN THIS POST HAS BEEN RETRACTED AND FOLLOW UP STUDIES HAVE FAILED TO REPEAT THE RESULTS DISCUSSED BELOW
Biologists are going gaga over the newest gene-editing protein - a DNA-cleaving Argonaute from Natronobacterium gregoryi, or NgAgo for short. Addgene has already distributed this plasmid all over the world, and the question on everyone’s minds is: could NgAgo replace CRISPR? Such a drastic shift won’t happen overnight, but there are a few reasons why you might choose NgAgo over CRISPR proteins Cas9 or Cpf1 - keep reading to learn more!
Background: Argonaute and NgAgo
In eukaryotes, Argonaute proteins play an important role in RNA interference, binding RNA guides to cleave foreign RNA. Argonaute proteins are also present in prokaryotes, and some of these proteins protect against invading DNA. In 2014, Swarts et al. characterized Argonaute from Thermus thermophilus (TtAgo), showing that this protein could cleave DNA in vitro, directed by phosphorylated DNA guides. Unfortunately, Swarts et al. were unable to show TtAgo-mediated cleavage in mammalian cells, likely due to high temperature requirements.
To solve this problem, Gao et al. searched for Argonaute orthologs similar to TtAgo and PfAgo, another thermophilic Argonaute. They identified NgAgo and showed that it can cleave plasmid and genomic DNA in a variety of mammalian cell lines, with an efficiency similar to that of Cas9. Like Cas9, NgAgo is also able to mediate homology-directed repair when a template is supplied.
Find Plasmids from the NgAgo Publication at Addgene
Cas9 and NgAgo head to head
NgAgo has a number of unique features compared to Cas9. These include: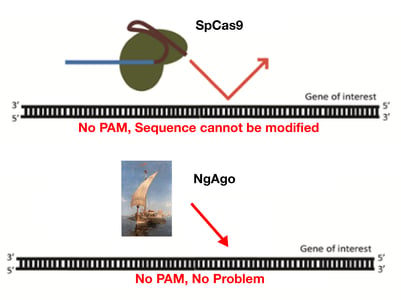
More targeting flexibility
Unlike Cas9 and the other CRISPR enzymes, NgAgo does not require a PAM sequence, so users should have much more flexibility in choosing a genomic target. NgAgo also appears to outperform Cas9 in GC-rich regions.
DNA guides
NgAgo uses 5’ phosphorylated DNA guides (so called “gDNAs”) rather than the RNA guides employed by Cas9 or Cpf1. These short, 24 base DNA guides may be more user-friendly, since they can be ordered as oligonucleotides and should be stable in the cell. gDNA guides will also need to be transfected into the cells of interest. In contrast, RNA guides must be expressed from a plasmid or in vitro transcribed, and changes in gRNA secondary structure can affect editing efficiency. Gao et al. tested if NgAgo preferred certain guide sequences - using 5 guides/gene for 10 genes, they did not observe differences in cleavage efficiency. If this finding is replicated in larger studies, it will represent a key advantage over Cas9, for which gRNA sequence can greatly affect editing efficiency.
Smaller size
At only 887 amino acids in length, NgAgo is shorter than SpCas9 (1368 aa), SaCas9 (1053 aa), or Cpf1 (1228 or 1307 aa depending on the species). This feature is advantageous if you’re working with small capacity viral vectors such as AAV.
Base removal
RNA-guided CRISPR enzymes cause double-stranded breaks, with either blunt (Cas9) or staggered (Cpf1) cleavage. NgAgo takes a different approach, randomly removing 1-20 nucleotides from the cleavage site specified by the gDNA. Depending on your application, this could be favorable or unfavorable. In a natural context, it’s hypothesized that base removal helps prevent the invading genome from “recovering” its original sequence. If you’re looking to disrupt gene function via indel formation, this property could be an advantage. However, repair after base excision would alter the target sequence and preclude future HDR, which could lower HDR efficiency.
Higher sensitivity to off-targets
More research is needed to compare CRISPR and NgAgo, but it seems that NgAgo has lower potential for off-target effects. In Gao et al.’s experiments, a single mismatch at any position of the DNA guide impaired NgAgo activity, with three mismatches abolishing activity.
One-guide faithful
NgAgo guide loading appears limited to the period when the protein is being synthesized, so NgAgo remains faithful to its guide DNA and cannot switch guides. While this fidelity would prevent other small nuclear DNAs from binding NgAgo, it might make it difficult to use NgAgo to disrupt multiple targets.
Are you using NgAgo? Let us know how it’s working in the comments section!
Read About NgAgo User's First Impressions Here
References
1. Gao, Feng, Xiao Z. Shen, Feng Jiang, Yongqiang Wu & Chunyu Han. “DNA-guided genome editing using the Natronobacterium gregoryi Argonaute.” Nat Biotechnol. Epub 2016 May 2. PubMed PMID: 27136078.
Find plasmids from this publication at Addgene.
2. Swarts, Daan C., et al. “DNA-guided DNA interference by a prokaryotic Argonaute.” Nature. 507(7491) (2014):258-61. PubMed PMID: 24531762. PubMed Central PMCID: PMC4697943.
- Find plasmids from this publication at Addgene.
3. Swarts, Daan C., et al. “Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA.” Nucleic Acids Res. 43(10) (2015):5120-9. PubMed PMID: 25925567. PubMed Central PMCID: PMC4446448.
Additional Resources on the Addgene Blog
- Read About Cas9 Homolog Cpf1
- Check Out High Fidelty Cas9 Mutants
- Browse All Genome Engineering Posts
Additional Resources on Addgene.org
- Check Out Other Argonaute Technologies from the Doudna Lab
- Browse the CRISPR Collection
- Find CRISPR Pooled Libraries
Topics: CRISPR, Cas Proteins





Leave a Comment