The ocean is full of striking visuals from fluorescent jellyfish, bioluminescent phytoplankton, and colorful corals, to name a few. What’s behind these sights are an abundance of biological molecules like chromoproteins, fluorescent proteins, and luminescent proteins. While fluorescent proteins and luminescence have been widely used in biological research for a long time, chromoproteins are just now being developed for a broad range of biological inquiries.
What is a chromoprotein and how is it different from fluorescent proteins?
Before going into the difference between chromoproteins and fluorescent proteins, it’s important to note that despite their distinction by name, chromoproteins are actually a subset of the fluorescent protein family. What sets them apart is the way they generate their color: they strongly absorb visible light to give colors in ambient light. In contrast, the widely used green fluorescent protein (GFP) gives a bright green fluorescence when exposed to blue to ultraviolet light, whereas chromoproteins do not fluoresce at all or fluoresce at very low levels.
Chromoproteins in nature
Compared to the history of fluorescent proteins in molecular biology research, chromoproteins have a shorter history. In 1987, scientists described an unusual blue-colored protein from the jellyfish Cassiopea xamachana (Blanquet and Phelan, 1987). Eight years later, scientists isolated pink, purple, and blue chromoproteins isolated from reef-building corals (Dove et al., 1995). In the early 2000s, a handful of chromoproteins were identified in sea anemone (Chan et al., 2006).
Why do these organisms have chromoproteins? One reason may be that chromoproteins protect corals from solar irradiation by reflecting visible and infrared light to dissipate excess energy at wavelengths of low photosynthetic activity. This still allows photosynthetically active wavelengths to reach zooxanthellae, the coral’s symbiotic dinoflagellates that are photosynthetic (Salih et al., 2000). Chromoproteins also enhance corals’ resistance to mass bleaching, the loss of their symbiotic algae, during heat stress. For other organisms, like the sea anemone, scientists think that chromoproteins could be used for mimicry and camouflage. An anemone’s red colored tentacles resemble worms which attract fish for them to prey upon (Figure 1).
 |
| Figure 1: The sea anemone's red color can be used for mimicry. Image: Bernard Spragg. |
Advantages of using chromoproteins for molecular biology
Because chromoproteins absorb visible light and give off color in ambient light, it gives scientists the ability for instrument-free detection. Unlike fluorescence or luminescence, which require UV lamps, fluorometers, or luminometers, chromoprotein detection can be done by the naked eye.
This makes cloning quite a bit easier since you can see if your cloning experiment worked just by looking at the cells on the plate. For example, if your plasmid contains a gene encoding a blue chromoprotein, after transforming them into your cells, you can simply look for blue colonies to determine which cells have taken up the plasmid. Of course, other follow up methods like sequencing are more definitive, but visual identification is a good place to start before taking on more time consuming and expensive methods.
While techniques such as blue-white screening require X-gal, an expensive exogenously-added substrate, chromoproteins don’t require other substrates for screening. Thus chromoproteins can be used as markers in cases where cost and resources are limited or for bioart and teaching applications.
Find a chromoprotein for your experiment!
Eukaryotic chromoproteins for bacterial synthetic biology
Because the common chromoproteins used in the lab come from eukaryotes, they can be difficult to directly use in bacteria, where most genetic manipulations start. Anthony Forster’s lab used codon optimization programs to engineer 14 chromoprotein sequences into E. coli BioBrick plasmids (meffRed, eforRed, asPink, spisPink, scOrange, fwYellow, amilGFP, amajLime, cjBlue, meffBlue, aeBlue, amilCP, tsPurple, and gfasPurple) (Figure 2).
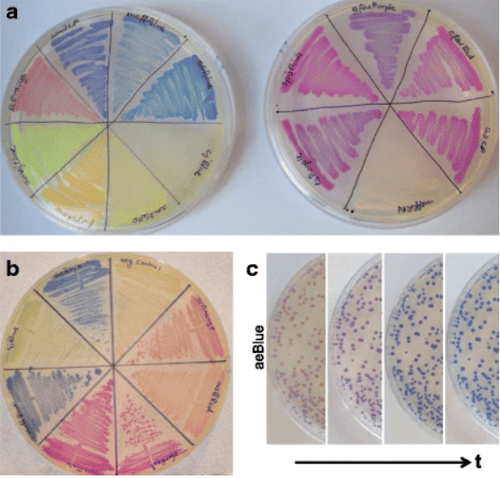 |
| Figure 2: Chromoprotein expression from bacteria on agar plate. Image: Liljeruhm et al., 2018. |
For each chromoprotein, the lab determined the color intensities, maturation times, and fitness cost of expression. They found that the cellular fitness cost varied greatly between chromoproteins. In some cases, cells containing high-copy plasmid-borne chromoproteins lost color overnight in liquid culture, though expressing the chromoproteins from the chromosome could get around this problem. There is no single “ideal” chromoprotein plasmid that combines the most desirable features: intense color, fast maturation, and low fitness cost yet. However, many fluorescent proteins face similar limitations, and many labs are already using these chromoproteins for specific purposes.
Chromoproteins for allelic exchange
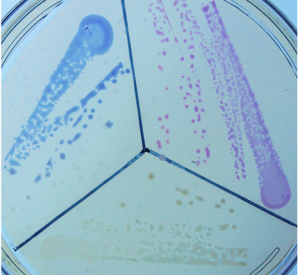 |
| Figure 3: pTOX plasmids expressing amilCP and tsPurple genes. Image: Lazarus et al., 2019. |
One such lab is the Waldor lab who used the amilCP and tsPurple chromoproteins to create negative selection markers for allelic exchange. These vectors contain a toxin gene (regulated by a rhamnose-inducible promoter) and a chromoprotein gene. Allelic exchange occurs through two sequential homologous recombination events:
- The plasmid carrying the desired genetic modification integrates into the genome (Figure 3). Here, the chromoprotein is expressed and gives the cell a magenta (amilCP) or purple (tsPurple) color. Though the toxin is integrated in the genome, it is not expressed as it is under the control of a rhamnose-inducible promoter.
- The resolution step leads to the desired mutation and excision of the vector backbone. To do this, rhamnose is added which induces expression of the toxin. Any cells that grow after this point will be those that have excised the vector backbone.
Vectors pTOX4, pTOX5, and pTOX6 express the YhaV, MqsR, and Tse2 toxins, respectively, and all encode the amilCP chromoprotein. Vectors pTOX7, pTOX8, and pTOX9 express the YhaV, MqsR, and Tse2 toxins, respectively, and encode the tsPurple chromoprotein.
Find the allelic exchange vectors here!
ShadowR, a novel FLIM-FRET acceptor
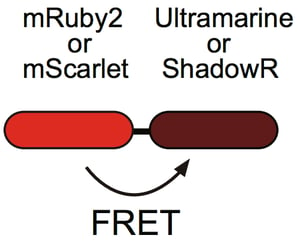 |
| Figure 4: ShadowR is used as a FRET acceptor to mRuby2 or mScarlet. Image: Murakoshi et al., 2019. |
ShadowR is an orange light-absorbing chromoprotein that Hideji Murakoshi’s lab developed as an acceptor for fluorescence lifetime imaging microscopy-based Förster resonance energy transfer (FLIM-FRET) in living cells (Figure 4). They created ShadowR by modifying the surface of the chromoprotein Ultramarine with hydrophilic amino acids to reduce non-specific interactions with cytosolic proteins. This step is important as non-specific binding to proteins can inhibit their activity. ShadowR can be used as a FRET acceptor in combination with mRuby2 or mScarlet as the donor in HeLa cells.
Find ShadowR for mammalian cell expression here
Now that you’ve learned what chromoproteins are and how they can be used, why not add some color into your research? If you have a favorite chromoprotein, either because it works really well for your experiment or you like how it looks for agar art, let us know!
Additional resources on the Addgene blog
- Find our Fluorescent Protein 101 blog series
- Learn more about FRET
- Find protocols and tips for making plasmid constructs
Resources on Addgene.org
- Find fluorescent proteins for your research
- Read our Molecular Biology Reference
- Find plasmids for FRET
References
Blanquet RS, Phelan MA (1987) An unusual blue mesogleal protein from the mangrove jellyfish Cassiopea xamachana. Marine Biology 94:423–430 . https://doi.org/10.1007/bf00428249
Chan MCY, Karasawa S, Mizuno H, Bosanac I, Ho D, Privé GG, Miyawaki A, Ikura M (2006) Structural Characterization of a Blue Chromoprotein and Its Yellow Mutant from the Sea AnemoneCnidopus Japonicus. Journal of Biological Chemistry 281:37813–37819 . https://doi.org/10.1074/jbc.m606921200
Dove SG, Takabayashi M, Hoegh-Guldberg O (1995) Isolation and Partial Characterization of the Pink and Blue Pigments of Pocilloporid and Acroporid Corals. The Biological Bulletin 189:288–297 . https://doi.org/10.2307/1542146
Lazarus, J. E., Warr, A. R., Kuehl, C. J., Giorgio, R. T., Davis, B. M., & Waldor, M. K. (2019). A New Suite of Allelic-Exchange Vectors for the Scarless Modification of Proteobacterial Genomes. Applied and Environmental Microbiology, 85(16). https://doi.org/10.1128/aem.00990-19
Liljeruhm, J., Funk, S. K., Tietscher, S., Edlund, A. D., Jamal, S., Wistrand-Yuen, P., … Forster, A. C. (2018). Engineering a palette of eukaryotic chromoproteins for bacterial synthetic biology. Journal of Biological Engineering, 12(1). https://doi.org/10.1186/s13036-018-0100-0
Murakoshi, H., Horiuchi, H., Kosugi, T., Onda, M., Sato, A., Koga, N., & Nabekura, J. (2019). ShadowR: a novel chromoprotein with reduced non-specific binding and improved expression in living cells. Scientific Reports, 9(1). https://doi.org/10.1038/s41598-019-48604-4
Salih A, Larkum A, Cox G, Kühl M, Hoegh-Guldberg O (2000) Fluorescent pigments in corals are photoprotective. Nature 408:850–853 . https://doi.org/10.1038/35048564
Topics: Fluorescent Proteins, Other Fluorescent Protein Tools







Leave a Comment