This post was contributed by guest bloggers Joachim Goedhart and Marieke Mastop from the Section of Molecular Cytology and Van Leeuwenhoek Centre for Advanced microscopy, University of Amsterdam.
The previous two posts in this series described a practical approach to selecting a bright fluorescent protein and a photostable fluorescent protein. In the third post of this series, we will discuss how to select a non-aggregating fluorescent protein.
In the jellyfish Aequorea victoria, AvGFP forms a homodimer. In corals, the red fluorescent proteins form tetramers. In general, fluorescent proteins have a natural affinity and a tendency to form higher order aggregates. This property can be tolerated in some applications (e.g. labeling of cells or tracking promotor activity), but it is problematic in applications in which the fluorescent protein is used as an inert protein module. This is explained in more detail here. There are a variety of methods that can be used to measure your fluorescent protein’s propensity to aggregate. The basics and pitfalls of these experiments are discussed here.

In vitro tests for aggregation
The tendency of purified fluorescent proteins to form homodimers or higher order oligomers can be analyzed by several techniques. Gel filtration and SDS-PAGE under non-denaturing conditions separate proteins based on size. Whether higher molecular weight complexes are detected will depend on the homodimer affinity and the concentration of the protein solution. During the experiment, the solution is diluted, complicating the interpretation. Because of this, these methods only give a qualitative view of dimerization tendency. Another technique that separates molecules based on size is ultracentrifugation. Sedimentation equilibrium analytical ultracentrifugation yields an affinity and has been used to demonstrate that yellow fluorescent protein homodimerization is characterized by an affinity of 110 µM (Zacharias et al, 2002), meaning that at this concentration 50% of the protein forms dimers. Finally, spectroscopic methods (based on fluorescence polarization) can be used to detect homodimerization. However, all the aforementioned methods only determine the capacity of homodimerization in aqueous solution.
In cyto tests for aggregation
 To what extent does homodimerization assayed with purified protein, reflect the likelihood that fluorescent proteins interact or oligomerize in cells? This question is largely unanswered. Ideally, fluorescent proteins should act as inert modules that fail to homodimerize in cells. To understand how well fluorescent proteins approach this ideal situation, cellular assays have been proposed. Early efforts tested whether or not FP-tubulin fusion proteins could properly form microtubule networks in cells. Other efforts tested whether FP fusions to connexin localized correctly to gap junctions (Shaner et al, 2008). Obligate dimers (such as dTomato) that promote interactions between the FPs interfere with native interactions, preventing proper localization in these assays. However, these strategies will not detect weak dimerization tendency, as is present in EGFP.
To what extent does homodimerization assayed with purified protein, reflect the likelihood that fluorescent proteins interact or oligomerize in cells? This question is largely unanswered. Ideally, fluorescent proteins should act as inert modules that fail to homodimerize in cells. To understand how well fluorescent proteins approach this ideal situation, cellular assays have been proposed. Early efforts tested whether or not FP-tubulin fusion proteins could properly form microtubule networks in cells. Other efforts tested whether FP fusions to connexin localized correctly to gap junctions (Shaner et al, 2008). Obligate dimers (such as dTomato) that promote interactions between the FPs interfere with native interactions, preventing proper localization in these assays. However, these strategies will not detect weak dimerization tendency, as is present in EGFP.
Constantini et al, (2012) developed a cell-based assay that does a better job of detecting homodimerization tendency. This assay employs a peptide derived from cytochrome p450, CytERM, to direct the fluorescent protein to the Endoplasmic Reticulum (ER). If homodimerization occurs (most likely in an antiparallel configuration), typical structures known as Organized Smooth Endoplasmic Reticulum (OSER) whorls are visible. These whorls can be quantified for individual cells, and the number of 'normal' looking cells can be counted. This assay will provide information on the dimerization tendency in living cells. Strikingly, the OSER assay reveals that tagRFP does not act as a true monomer in cells (Constantini, 2012), whereas it was previously concluded to be monomeric based on gel filtration (Chudakov et al, 2007).
We have developed a variety of plasmids that can be used in the OSER assay including CytERM-mKOkappa, CytERM-mTurquoise2, and CytERM-dTomato.
When using and interpreting the results of the OSER assay, you should keep in mind that the presence of whorls is probably concentration dependent and cell-type dependent. Therefore, it is good practice to perform this assay with your chosen FP in your cell type of interest before starting your experiments.
Selecting a non-oligomerizing protein
A fluorescent protein cannot be assumed to be non-dimerizing if its name is preceded by an 'm' to declare that it is monomeric. The claim that a fluorescent protein is monomeric without the accompanying data to prove it is worthless. The OSER assay is currently the best way to test a fluorescent protein’s propensity to homodimerize. Several studies have used the OSER assay to demonstrate monomeric behaviour in cells (Shaner et al, 2013; Bindels et al, 2017).
Still, the data presented in papers should be used with caution, since the difference between studies is substantial (e.g, the poor performance of mRuby2 is detected by Constantini et al (2015) and by Bindels et al (2017) but not by Cranfill et al (2016)). Moreover, good performance in the OSER assay is not a guarantee that using this fluorescent protein to tag your protein of interest will be problem-free.
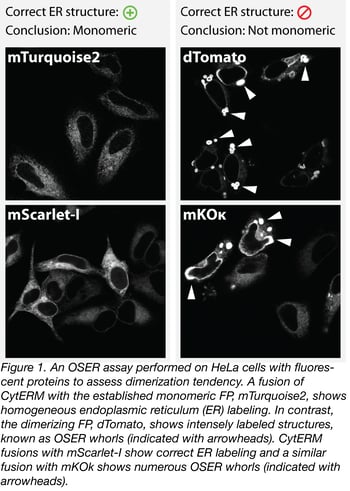
In figure 1 we provide an example of the OSER assay with a number of fluorescent proteins. Fusions of CytERM with mTurquoise2 and dTomato, which are established monomeric and dimerizing FPs respectively, are used as positive and negative controls for proper ER structure. Based on these data, we conclude that mKOk has dimerization tendency and is not suitable for protein tagging. On the other hand, mScarlet-I shows correct ER labeling, which is good evidence that mScarlet-I acts as a monomer in the cell type tested.
To conclude, we recommend that you try a couple of different fluorescent proteins when protein tagging is the goal (Cranfill et al, 2016; Constantini et al, 2012). The localization and biological properties of new fusions can be compared with mEGFP or mTurquoise2, which are established true monomeric fluorescent proteins.
Concluding remarks
Many fluorescent proteins are available, and their number is steadily increasing. Of course, you want the ‘latest and greatest’ variants with the best properties for your research. But be aware that the fluorescent proteins reported in publications are only characterized under a limited number of conditions, often using purified proteins. Therefore, it is essential to verify a number of key properties (brightness, photostability, oligomerization) of a number of promising fluorescent proteins under the conditions that are relevant for your research.
This will reveal the performance of fluorescent proteins in your biological system with the microscope that you will be using. If you do a head-to-head comparison right, it will be valuable information for the scientific community. The knowledge and experience that is gathered can be shared by publishing it as (part of) figure 1, uploading it as a preprint, or documenting it in some other way. Besides the data, new plasmids and/or cells or organisms that are engineered can be shared to simplify the comparison by others. Together, we can build a valuable resource with tools and data that indicate the performance of (a set of) fluorescent proteins under specific conditions.
Many thanks to our guest bloggers Joachim Goedhart and Marieke Mastop!
 Joachim Goedhart is an assistant professor at the Section of Molecular Cytology and Van Leeuwenhoek Centre for Advanced microscopy (University of Amsterdam). He develops, characterizes and uses genetically encoded fluorescent probes. You can follow him on twitter: @joachimgoedhart.
Joachim Goedhart is an assistant professor at the Section of Molecular Cytology and Van Leeuwenhoek Centre for Advanced microscopy (University of Amsterdam). He develops, characterizes and uses genetically encoded fluorescent probes. You can follow him on twitter: @joachimgoedhart.
 Marieke Mastop is a PhD candidate at the Section of Molecular Cytology (University of Amsterdam) where she develops genetically encoded FRET based biosensors.
Marieke Mastop is a PhD candidate at the Section of Molecular Cytology (University of Amsterdam) where she develops genetically encoded FRET based biosensors.
Additional Resources on the Addgene Blog
- Which Fluorescent Protein Should I Use?
- Choosing Your Fluorescent Proteins for Multicolor Imaging
- Learn about Fluorescent Protein Biosensors
Additional Resources on Addgene.org
- Find Empty Backbones with Fluorescent Tags
- Find Plasmids for Subcellular Localization
- Browse the entire Fluorescent Protein Collection





Leave a Comment