This post was contributed by Patrick Miller-Rhodes from the University of Rochester Medical Center.
You’ve probably heard of Forster Resonance Energy Transfer (FRET). Through the non-radiative transfer for energy between neighboring fluorophores, FRET can be used to detect the inter- and intramolecular interactions that underlie protein function. However, FRET experiments can be difficult to implement in practice because FRET depends on a number of hard-to-achieve factors. For example, FRET requires that fusion proteins be in close proximity and present in large enough quantities (and the correct stoichiometric ratios) to generate useable data. What’s more, measuring and quantifying FRET is often easier said than done.
Fortunately, a complementary method exists for visualizing protein-protein interactions (PPIs): Bimolecular Fluorescence Complementation (BiFC).
What is bimolecular fluorescence complementation?
Bimolecular fluorescence complementation is a method for detecting protein-protein interactions that uses split fluorescent proteins tethered to potential protein interaction partners. Unlike FRET, BiFC only uses a single fluorescent protein to visualize PPIs. This fluorescent protein (FP) is split into two fragments that individually lack fluorescence. When complementary FP fragments are fused to two hypothesized protein interaction partners (PIPs), the FP fragments form the complete FP because the PIPs they’re tethered to bring them into close proximity (Figure 1). Formation of the complete FP results in a fluorescent signal that can be detected by fluorescence microscopy.
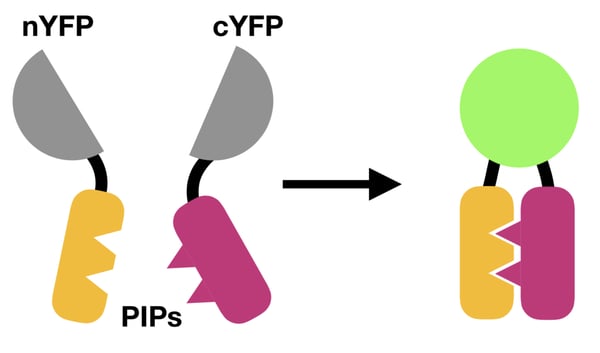 |
| Figure 1: How BiFC works. A split FP (e.g., yellow fluorescent protein, YFP) is reunited by the interaction between two PIPs, resulting in a fluorescent readout. |
Applications for bimolecular fluorescence complementation
BiFC has been used to directly visualize PPIs that regulate numerous cellular functions, including ubiquitinoylation, kinase signaling, integrin signaling, and transcription factor interactions (Kerppola, 2006). One of the first BiFC studies used the method to interrogate the subcellular localization of interactions between basic leucine zipper and Rel transcription factors (Hu et al., 2002). This would be more difficult with FRET, as the overexpression of fusion proteins it requires can lead to aberrant subcellular localization of the studied PIPs.
The straightforward nature of BiFC also makes it easier to generate a large number of constructs for high-throughput screening of PPIs. For example, Bischof et al. (2018) generated an open reading frame (ORF) library to screen PPIs among 450 transcription factors in live fruit flies. Although BiFC open-reading frame (ORF) libraries have been generated for yeast (Kim et al., 2019) and fruit flies (Bischof et al., 2018), the generation of such resources in less genetically tractable organisms lags behind.
BiFC can also be used to screen for small-molecule inhibitors of a particular PPI. In such experiments, small-molecule inhibitors are tested in parallel for their ability to prevent BiFC resulting from the assayed PPI. This high-throughput application is much more straightforward to carry out because it relies on a single BiFC pair.
Find BiFC plasmids for your experiment at Addgene!
How to Design a BiFC Experiment
Much like FRET, BiFC experiments must be carefully optimized to be successful. A BiFC experiment generally involves three steps: fusion protein construction, cellular expression, and quantification (Kerppola, 2006, 2013). These are described in more detail below:
Fusion protein construction
Fusing FP fragments to your PIPs requires careful selection of FP fragments and, preferably, some information about the structures of the tested PIPs. Many split FPs have been validated for BiFC (Kodama and Hu, 2012). Having a rough idea of how your PIPs might interact will allow you to determine where to fuse the FP fragments. In the absence of this information, you can fuse each FP fragment to the N- and C-terminal end of each PIP and empirically test which combination provides the best fluorescent signal. As for linker sequences, the amino acid sequences RSIAT and RPACKIPNDLKQKVMNH have been used successfully (Kerppola, 2006). Flexible GS linkers may also work. You will also need to generate negative control constructs — more on this later. For more on constructing fusion proteins, check out our article: Fluorescent Proteins 101: GFP Fusion Proteins - Making the Right Connection.
Cellular expression
Whether BiFC constructs are expressed transiently by transfection or stably impacts the success of BiFC experiments. In either case, overexpression should be avoided because it can generate high background fluorescence levels due to the random association between FP fragments independent of PPIs. Overexpression can be assessed by comparing the expression levels between BiFC constructs and their endogenous counterparts using western blotting. The subcellular localization of BiFC constructs should also be compared against those of their endogenous counterparts (by immunocytochemistry, for example), as this can also be perturbed by supraphysiological expression levels.
BiFC quantification
This is where BiFC outshines FRET — BiFC experiments only require that you image and measure the fluorescence of a single FP. Your control construct may generate a modest level of fluorescence due to the random association between FPs independent of PPIs. Simply compare the fluorescence intensity of your BiFC constructs to your negative control to determine whether your PIPs actually do interact.
Choosing the right negative control is critical to the success of BiFC experiments. The ideal control is a mutated variant of one or both of your PIPs in which the protein-interaction interface has been compromised (Kodama and Hu, 2012). Such controls are necessary because split FPs can associate, albeit at relatively low frequencies, without being brought into close proximity by PIPs. The FP fragments themselves cannot be used as negative controls because their expression level, stability, and subcellular localization may differ from those of your BiFC construct, making it difficult to interpret your results. Unfortunately, without structural information, constructing appropriate controls may requires that you empirically test a number of mutants to identify those that inhibit BiFC in your cellular system.
BiFC competition analysis can be used in lieu of negative controls (Kerppola, 2006). Here, one of the endogenous PIPs is expressed at increasing levels alongside your BiFC constructs. If your PIPs actually interact, co-expressing one of the endogenous PIPs should decrease the BiFC signal in a dose-dependent manner. This is straightforward in easy-to-transfect cell lines where plasmid dosage can be tightly controlled. This is also theoretically possible, but likely more challenging, with lentiviral expression.
For more detail on how to design BiFC experiments, detailed troubleshooting tips can be found in protocols by Kerppola (2006, 2013).
BiFC vs. FRET: When to choose one over the other
BiFC is a convenient, fluorescence-based method for directly visualizing PPIs in cells. The ease with which BiFC can be visualized makes it amenable to studying PPIs under relatively physiological conditions when compared to FRET. However, it’s important to note that BiFC is irreversible — FP fragments become covalently bonded by associating with one another — which means it cannot be used to measure the dissociation of PIPs. Thus, whereas BiFC is useful in scenarios that call for a simple, yes-no answer — i.e., do two proteins interact or not? — FRET is preferable when the temporal dynamics of PPIs are the focus of study.
Many thanks to our guest blogger Patrick Miller-Rhodes from the University of Rochester Medical Center!

Patrick is equal parts neuroscientist, writer, and extreme sports enthusiast. He recently obtained his Ph.D. in neuroscience from the University of Rochester Medical Center, where he used molecular biology techniques to study inflammation and neurodegeneration. He now spends his time learning and writing about science whenever he's not off the grid.
References
Bischof J, Duffraisse M, Furger E, Ajuria L, Giraud G, Vanderperre S, Paul R, Björklund M, Ahr D, Ahmed AW, Spinelli L, Brun C, Basler K, Merabet S (2018) Generation of a versatile BiFC ORFeome library for analyzing protein–protein interactions in live Drosophila. eLife 7. https://doi.org/10.7554/elife.38853
Hu C-D, Chinenov Y, Kerppola TK (2002) Visualization of Interactions among bZIP and Rel Family Proteins in Living Cells Using Bimolecular Fluorescence Complementation. Molecular Cell 9:789–798. https://doi.org/10.1016/s1097-2765(02)00496-3
Kerppola TK (2006) Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc 1:1278–1286. https://doi.org/10.1038/nprot.2006.201
Kerppola TK (2013) Bimolecular Fluorescence Complementation (BiFC) Analysis of Protein Interactions in Live Cells. Cold Spring Harb Protoc 2013:pdb.prot076497. https://doi.org/10.1101/pdb.prot076497
Kim Y, Jung JP, Pack C-G, Huh W-K (2018) Global analysis of protein homomerization in Saccharomyces cerevisiae. Genome Res 29:135–145. https://doi.org/10.1101/gr.231860.117
Kodama Y, Hu C-D (2012) Bimolecular fluorescence complementation (BiFC): A 5-year update and future perspectives. Biotech 53. https://doi.org/10.2144/000113943
Additional resources on the Addgene blog
- Browse our Fluorescent Protein 101 blog posts
- Read our blog post covering an introduction to FRET
Resources on Addgene.org
Topics: Fluorescent Proteins, FRET






Leave a Comment