 This post was contributed by guest blogger, Jennifer Rivers, a research microbiologist specializing in bacteriology and drug resistance.
This post was contributed by guest blogger, Jennifer Rivers, a research microbiologist specializing in bacteriology and drug resistance.
It seems as though, nowadays, we cannot make it a full month without a new, drug-resistant ‘superbug’ making headlines in one part of the world or another. Bacterial resistance to antibiotics is certainly a topic of great concern in healthcare today, but it doesn’t have to be some looming, abstract, convoluted scientific concept. It is imperative that we all understand what antibiotic resistance is and how it develops so that we may take an active part in our own health and the health of our loved ones, and become a positive force for public health.
Visit Addgene's Microbiology Pages
What is antibiotic resistance?
Antibiotic resistance is defined as microbial survival despite exposure to antibiotics designed to kill them or to impede their growth. When we hear the word ‘antibiotic’, very often we immediately imagine a prescription for a bacterial infection. While this post will, indeed, focus on bacteria, it is important to note that the term ‘antibiotic’ can also be used to describe anti-parasitic drugs and anti-fungal drugs, because ‘antibiotic’ merely means that the drug prevents the growth of a living organism. As for multi-drug resistance: definitions vary in the literature, but generally organisms with multi-drug resistance are resistant to three or more different classes, or types, of drugs used to treat them. Very often, this is what it means to be a ‘superbug’. Superbugs are often very difficult and very time consuming to treat.
Learn How Antibiotic Resistance Genes Are Used in the Lab
How does antibiotic resistance develop?
There are many ways in which bacteria can become resistant to antibacterial drugs. Resistance can be intrinsic, meaning a basic characteristic that naturally occurs in the bacterium, and is essentially always there, or acquired, which means the bacterium is not naturally resistant but has become resistant somehow overtime. One fantastic example of intrinsic resistance would be Mycoplasma species’ resistance to beta-lactam antibiotics, like penicillin. These specific drugs attack the cell wall of bacterial cells; however, Mycoplasma species do not possess cell walls. As a result, Mycoplasma are intrinsically resistant to this class of antibiotic. Acquired resistance can be the result of one bacterium sharing genes with another, an accidental transmission of genes from one bacterium to another through bacteriophage (viruses that only infect bacterial cells), or a bacterium picking up resistance genes from it’s environment (from DNA left behind after another bacterium died for instance). These three mechanisms of acquired drug resistance are referred to as methods of ‘horizontal gene transfer’. Resistance can also arise from random mutations in the genetic sequence that happen to be beneficial, and persist as a result.
Why does antibiotic resistance develop?
Microbes (bacteria included) are very skilled at the delicate art of survival. Existing virtually ubiquitously in the environment, there is an intense amount of competition between microorganisms for available resources. This competition drives microbes to develop survival mechanisms. Some microbes even make their own antibiotic compounds. In such a case, the microbe itself would be unharmed by the compound it produces (think intrinsic resistance), but surrounding microbes would be affected (possibly killed), thus increasing the initial microbe’s chances of survival. This was the whole basis of how penicillin was discovered! Of course we wouldn’t have such a rich diversity of microbes in the world if some didn’t develop, either through random mutation or horizontal gene transfer, means to defend themselves from these naturally occurring antibiotics. When we use antibiotics to treat infections, we are treating microbes with compounds similar to those they have been battling for centuries. As such, resistant populations can arise very quickly following the tried and true process of evolution.
Watch this video showing bacteria evolve antibiotic resistance on a plate!
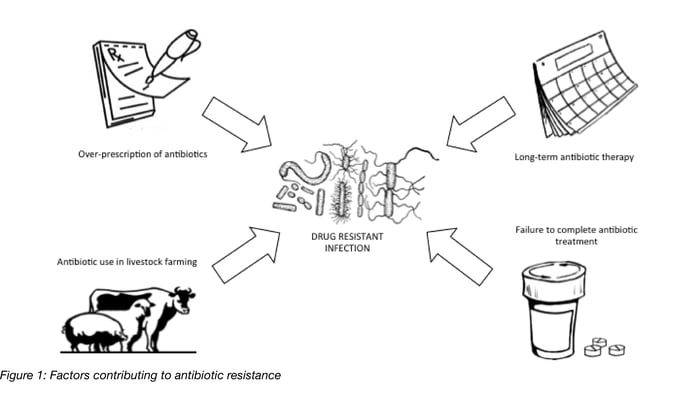
Antibiotic resistance can arise, and has arisen, for many different reasons. Exposure to antibacterial drugs provides a selection pressure, killing off bacteria that are susceptible to the drug, while the few that happen to possess resistance genes survive and multiply. Factors contributing to the development of resistance include (Figure 1):
- Over-prescription of antibiotics (for example, giving antibiotics for an infection that isn’t caused by bacteria, like the cold or flu)
- Overuse of antibiotics in livestock farming
- Lengthy courses of treatment with antibiotics
These scenarios expose bacterial populations to antibiotics at unnecessary times, putting pressure on resistant bacteria to over-take the population and making them more difficult to treat in the future.
- Patients not finishing the whole course of antibiotics prescribed to them
This may seem counter-intuitive considering the other three scenarios, but a short treatment regiment might wipe out bacteria that are completely susceptible to a drug while a full course of treatment is required to wipe out those bacteria that are partially resistant. If you stop a treatment early, you may be un-wittingly selecting for less susceptible bacteria that will be more difficult to treat later. A similar situation can occur if you treat a bacterial infection with a lower than optimal drug concentration. Drug resistance is not a black or white issue. Sometimes bacteria are resistant to lower concentrations of a drug, but will be killed by a higher concentration, or sometimes more exposure time is needed for the drug to work… if the instructions for treatment aren’t followed closely, you could be doing more harm than good, even with the best of intentions. The take-home? Always take your medication as prescribed.
How do we know which bacteria are resistant to antibiotics and which ones aren't?
This video shows how to perform a Kirby-Bauer disk diffusion test and check bacteria for antibiotic resistance. Source: https://www.youtube.com/watch?v=sx1uDYSfINA.
When you go to the doctor, it will very likely take time to determine if you have an antibiotic resistant infection or not, because scientists in the laboratory will need to isolate and grow whatever bacteria are causing the infection before they can test for resistance. Growing and isolating the infecting bacteria can take 24 hours or more, and testing for resistance often takes an additional 18-24 hours, or even longer depending on the bacterial species. Because of this and in an effort to improve the health of the patient as quickly as possible, especially if the infection is serious or life-threatening, treatment will often begin before this information becomes available. For routine infections, like strep throat, testing for antibiotic resistance isn’t commonly performed because resistance in these infections is so rare.
When antibiotic resistance testing is requested clinically, this involves growing a standardized amount of the bacteria in the presence of a known concentration of antibiotic that has been impregnated into a paper disk, and observing whether or not bacterial growth is inhibited. The amount of inhibition around the disk is measured in millimeters, and corresponds to the level of susceptibility you might expect if a certain antibiotic were chosen for treatment. This is called a Kirby-Bauer disk diffusion test or Kirby-Bauer susceptibility test (Figure 2), and requires at least 18-24 hours to obtain results. While this is not the only method of determining drug resistance status, it is a very common, well-trusted method that is frequently used in the clinical laboratory.
What can we do to stop the emergence of antibiotic resistance?
The problem with this question is, unless an antibiotic targets an essential, conserved, non-negotiable process within the bacterial cell, antibiotic resistance is very likely to develop over time. It isn’t feasible to completely eliminate the development of resistance; it’s a natural process of life and one which has allowed bacteria to evolve and survive for millennia. What we can do, however, is fight smarter, not harder. We can utilize tactics like rotating antibiotic treatments, so that exposure to each antibiotic is minimized, while the infection is still effectively treated. We can prescribe antibiotics for the shortest time period possible to effectively manage a patient infection. Limiting the use of antibiotics in animal farming is already being recognized as an important step, as you can’t walk into any supermarket now without seeing a promotion for antibiotic-free, farmed meats. We can focus on creating new antibiotic therapies, and investigate more effective drug targets (ones that are less likely to mutate or change in order for the bacterium to survive).
Check out our recent blog post on CRISPR antimicrobials to learn how scientists are using CRISPR to fight bacteria.
From a societal standpoint, we can all work together to try and prevent outbreaks of drug resistant bacteria within communities, schools, and households by observing proper cleanliness and personal hygiene standards. Handwashing is an essential component of preventing infection, as is cleaning and disinfecting frequently touched surfaces like tabletops, toys, gym equipment, etc. Virtually any preventive measure designed to help keep you from getting sick can be helpful in preventing the spread of antibiotic resistant bacteria from one person to the next. For more information and tips on preventing the spread of antibiotic resistant bacteria, check out this resource on preventing antibiotic resistance from the CDC. Antibiotic resistance can be scary to think about, but we can all do our part to learn all we can, and minimize both our risk and the risk to others. Your friends, family, and physicians will thank you!
Many thanks to our Guest Blogger, Jennifer Rivers!
 Jennifer Rivers ScM, MLS(ASCP)CM is a research microbiologist specializing in bacteriology and drug resistance. Additionally, she is passionate about health equity, science communication, and public health.
Jennifer Rivers ScM, MLS(ASCP)CM is a research microbiologist specializing in bacteriology and drug resistance. Additionally, she is passionate about health equity, science communication, and public health.
References
1. O'Neill, J. "Tackling drug-resistant infections globally: final report and recommendations." London: Wellcome Trust & HM Government (2016).
2. CDC Information Pages on Antibiotics
3. WHO Antimicrobial Resistance Fact Sheet
Additional Resources on the Addgene Blog
- Learn How Antibiotic Resistance Genes Are Used in the Lab
- Probe Phage Genomes for Host Binding Proteins
- Learn How to Use Our Microbiology Pages
Additional Resouces on Addgene.org
- Check Out Our Microbiology Pages
Topics: Microbiology, Other






Leave a Comment